- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
FRAP Analysis of LET-23::GFP in the Vulval Epithelial Cells of Living Caenorhabditis elegans Larvae
Published: Vol 5, Iss 11, Jun 5, 2015 DOI: 10.21769/BioProtoc.1489 Views: 9203
Reviewed by: Peichuan ZhangFanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Monitoring the Recruitment and Fusion of Autophagosomes to Phagosomes During the Clearance of Apoptotic Cells in the Nematode Caenorhabditis elegans
Omar Peña-Ramos and Zheng Zhou
Nov 20, 2022 2136 Views

Preparation of Caenorhabditis elegans for Scoring of Muscle-derived Exophers
Katarzyna Banasiak [...] Wojciech Pokrzywa
Jan 5, 2023 1877 Views

SunTag-Based Single-Molecule Translation Imaging in Caenorhabditis elegans
Elise van der Salm [...] Suzan Ruijtenberg
Oct 20, 2025 2295 Views
Abstract
The Caenorhabditis elegans (C. elegans) vulva is a well-established system to study organ development as the molecular mechanisms that govern its formation are conserved in animals. Of special interest is the EGFR/RAS/MAPK signaling pathway that is required for fate acquisition and morphogenesis of the vulva. let-23 encodes the sole homologue of the epidermal growth factor receptor (EGFR), is expressed at the plasma membrane of the vulval precursor cells (VPCs) and is activated by LIN-3 EGF at the end of the L3 larval stage to initiate vulva development. LET-23 activity can be modulated through altering its subcellular and plasma membrane localization. To study the trafficking of EGF receptor in a living organism, we created a functional LET-23::GFP translational reporter worm line (Haag et al., 2014) and quantified the mobile fraction of LET-23::GFP at the basolateral membrane of the VPCs by fluorescence recovery after photobleaching (FRAP). Here we describe the protocol for LET-23::GFP FRAP at the basolateral membrane of the VPCs and the data analysis using FIJI (ImageJ).
Materials and Reagents
- C. elegans strain zhIs038 [let-23::gfp; unc-119(+)] (Haag et al., 2014)
Note: For information on maintenance of C. elegans: http://www.wormbook.org/chapters/www_strainmaintain/strainmaintain.html. - 4% agarose pads (for an example see http://www.wormatlas.org/agarpad.html by Monica Driscoll)
- M9 solution with 20 mM tetramisole hydrochloride (anesthetic)
- M9 buffer for C. elegans (see Recipes)
Equipment
- Standard microscopy slides and 18 x 18 cover slips
- Zeiss LSM710 confocal microscope equipped with 458/488/514 nm argon laser
- Computer with FIJI (ImageJ. National Institutes of Health) and appropriate Plugins for FRAP analysis (see below)
Software
- LSM710 ZEN software (ZEISS)
- FIJI (Image J) (National Institutes of Health)
Procedure
This protocol describes FRAP of LET-23::GFP at the basal membrane of the VPCs, thus the “bleach region” is a part of the basal membrane. The protocol may be applied to lateral and apical membranes as well.
- Microscopy
- Prepare an agarose pad using 4% melted agarose. Cut the edges of the pad to obtain a square of around 1 cm x 1 cm (Figure 1a). The agarose square should be smaller than the area of the cover slip (18 mm x 18 mm). Add 3 μl of 20 mM tetramisol and pick 20-30 worms from late L3 stage into the tetramisol drop. Carefully place the coverslip on the top of the pad and add M9 buffer with a pipette to fill in the space between the agar and the borders of the cover slip. Under these conditions animals survive for more than 1 h, but do not analyze animals that have been left more than 45 min on the slide.
Notes:- It is not possible to identify worms at the 4-cell stage (Pn.pxx stage) with the dissection scope. Pick several worms and confirm the 4-cell stage on the confocal microscope using weak fluorescence light to avoid photobleaching.
- Animals at the 4-cell stage of vulval development survive longer under the confocal scope.
- It is fundamental that the animals are not moving on the slide. Weak horizontal or vertical drifts can be digitally corrected (see below), but animals that move too much during the acquisition cannot be analyzed.
- It is not possible to identify worms at the 4-cell stage (Pn.pxx stage) with the dissection scope. Pick several worms and confirm the 4-cell stage on the confocal microscope using weak fluorescence light to avoid photobleaching.
- On the confocal scope: Identify the ventral region of the animal where the vulval cells are located under DIC (differential interference contrast) or transmission light. Use the 63x /1.4 NA oil lens and change to UV fluorescence to identify the VPCs expressing LET-23::GFP. Center the field in the VPCs.
- In the Zeiss LSM710 ZEN software, use the “Channel color lookup table” to correct the saturation for each animal using the “Master Gain” function.
- Crop/Zoom 4x and rotate image to place the VPC horizontally in the viewing field. It’s important to keep the 4x zoom throughout all experiments.
- Perform a “Snap image” (Figure 1b).
- Bleaching settings:
- On the “Bleaching” window select the 488 nm argon laser with 85% laser power.
- On the “Start bleaching after # scan” option select “2”, and in the “iterations” options select “5” times bleach repeat.
- On the “Regions” window click on “Acquisition” for region #1 and “Bleach” for region #2. Select with the “rectangle” function from the “Regions” window the four VPCs and define this region as “Acquisition” and “Analysis” (Figure 1b).
- Select an area of the basal or lateral membrane to be photo bleached with the “rectangle” function and define this region as “Bleach” and “Analysis” (Figure 1b).
- Under these settings approximately 70% of the signal is photo bleached. Record the fluorescence recovery for 296 sec, taking a frame every 8 sec (“Time Series” window) with a 488 nm laser excitation, at 20% power intensity, a pinhole equivalent to 2.12 Airey units, a frame size of 256 x 256 dpi and speed (Pixel Dwell) of 3.15 microsecond (Figure 1 b-c).
Note: Perform a few bleach experiments and analyze the data (see below) to determine if photo bleach and recovery occurs under these conditions. If photo bleach is not efficient, avoid increasing the percentage of laser power; instead increase the iterations to bleach several times repeatedly. If recovery does not occur, check for integrity of the animals and reduce the time they are mounted on the slide.
- On the “Bleaching” window select the 488 nm argon laser with 85% laser power.
- Save the experiment in the .lsm format.
- Prepare an agarose pad using 4% melted agarose. Cut the edges of the pad to obtain a square of around 1 cm x 1 cm (Figure 1a). The agarose square should be smaller than the area of the cover slip (18 mm x 18 mm). Add 3 μl of 20 mM tetramisol and pick 20-30 worms from late L3 stage into the tetramisol drop. Carefully place the coverslip on the top of the pad and add M9 buffer with a pipette to fill in the space between the agar and the borders of the cover slip. Under these conditions animals survive for more than 1 h, but do not analyze animals that have been left more than 45 min on the slide.
- Data handling and analysis
In FIJI (ImageJ)- Install the following Plug-Ins:
StackReg: http://bigwww.epfl.ch/thevenaz/stackreg/ (written by Philippe Thévenaz)
TurboReg: http://bigwww.epfl.ch/thevenaz/turboreg/ (written by Philippe Thévenaz)
Edge Detection:
http://imagejdocu.tudor.lu/doku.php?id=plugin:filter:edge_detection:start (written by Thomas Boudier and repackaged by Joris Meys)
FRAP Norm:
http://imagejdocu.tudor.lu/doku.php?id=plugin:analysis:frap_normalization:start (written by Joris Meys) (Phair et al., 2004)
The “StackReg” and “TurboReg” are used to align the images as worms usually move a little during the recording. - Open the .lsm files in FIJI and perform the following:
Plugins > StackReg > Transformation > Rigid body > ok
Plugins > FRAP Analysis > FRAP Norm
Two windows appear: “Pre Bleach” and “Post Bleach” Window (ignore these windows).
In Window FRAP Analysis > Settings change the following: Pre Bleaching slice: 2 and Post Bleaching Slice: 3. Time units between slides: 8 (sec) > ok.
Note: In our setup the first picture is the “snapshot”. Therefore the Pre Bleached slice is “2” and the Post bleach is “3”.
In Frap Analysis window: > Measurements > Automatic (Figure 1d). - Select the rectangle selection tool.
- Go to the image window and select the post bleach image.
- Select the FRAP region with the rectangle selection tool. Choose a rectangle size that covers all bleached area (Figure 1e). Use the pre-bleach and post-bleach pictures as reference.
- In the FRAP Analysis window > FRAP Region> Set ROI > Apply to Image > Measure.
A “Measurement Window” displays values of the images over time (this information will not be used, but keep it open).
With the rectangle tool, select a part of the picture outside of the fluorescent area (Figure 1e’) (Background) and repeat the process in the FRAP Analysis window, this time as: Background > Set ROI > Apply to Image > Measure (Figure 1d).
With the rectangle tool select the 4 cells (Figure 1e’’) and in FRAP Analysis window click on:
Whole Cell > Set ROI > Apply to Image > Measure (Figure 1d).
Go back to the Fluorescence image and select with the rectangle tool a region of the membrane that was not bleached. For example, the basal membrane of the cell adjacent to the bleached cell (Figure 1e’’’). In FRAP Analysis window repeat the above process for > Reference > Set ROI > Apply to Image > Measure. - Click “Normalize” to obtain the normalization results.
- This procedure is used to doubly normalize the data: with the Reference region (nonbleached membrane) and with the total fluorescence of the 4 VPCs in the single section.
- Copy all results to a data analysis software (e.g. Excel) and keep the columns “Time” and
- “Normalized” for the analysis. Plot the data: Fluorescence intensity (normalized) vs. time (sec) (Figure 1f).
- The plots can be used to estimate the immobile and mobile fractions of the analyzed molecule. The mobile fraction is the fraction of fluorescence that is recovered and is represented by the difference between the initial and final fluorescence. FRAP-bleached molecules that are immobile do not permit mobilization of incoming proteins into those sites at the membrane and represent the immobile fraction. For a comprehensive explanation see White and Stelzer (1999).
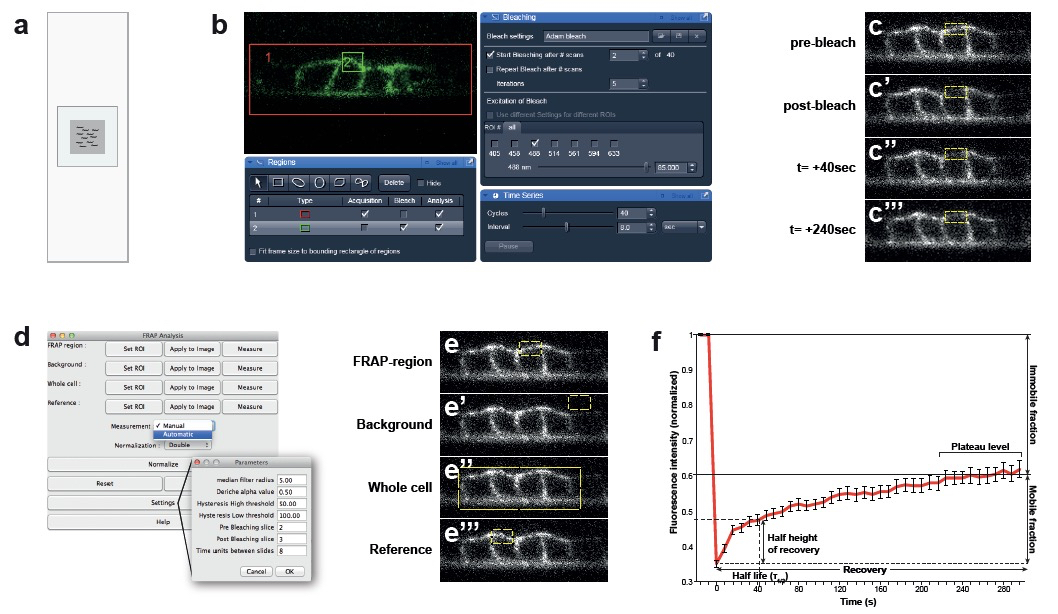
Figure 1. Confocal set up for bleach and recovery measurements and data analysis. a. Set up of the agar pad on the microscopy slide. For optimal immobilization of the animal, use agar pads that are smaller than the cover slip with 3 µl of tetramisole. b. Screenshots of the ZEN software. Bleach and recovering settings: Red rectangle “1” is the acquisition area (VPCs). Green rectangle “2” is the bleaching area (basal membrane). c. Images acquired before and after photobleaching of the basal membrane of the VPCs over 240 sec. d. Reference settings for the FRAP analysis plugin from FIJI. e. Selection of areas to normalize in the FRAP analysis plugin. f. Example of a FRAP curve of LET-23::GFP and its parameters (n= 40 animals).
- Go to the image window and select the post bleach image.
- Install the following Plug-Ins:
Notes
- FRAP approximately 30 animals per experiment. In some cases the animals move too much or die during the experiment. Some of the worms may not recover or stop recovering in the first minute. Before each measurement briefly determine the morphological integrity of the animal. Animals that have been left too long on the pad look less turgid and accumulate cavity-like structures in the head. Some worms may also “move” in the z-section as they collapse. Do not include these data for the analysis. We end up with around 20 animals that we use for the measurements.
- Only after the data is normalized and plotted it is possible to give an estimate of the recovery of the bleached protein. Before performing large experiments, perform a couple of pilot photo-bleaches and post-bleach acquisitions to determine if the settings are appropriate or if the animals are still alive under these conditions.
- Do not compromise speed for resolution: Speed of image acquisition is crucial for the FRAP analysis.
Microscope lens: Bleaching and acquisition with the 63x /1.4 NA oil lens give reproducible results in wild-type animals expressing LET-23::GFP. It might be necessary to use the 40x lens to collect more signals for mutant backgrounds or chemical treatments where LET-23::GPF expression may reduce.
Recipes
- M9 buffer for C. elegans
3.0 g KH2PO4
6.0 g Na2HPO4
0.5 g NaCl
1.0 g NH4Cl
Bring to 1 L with H2O
Acknowledgments
Funding: This work was supported by a grant from the Swiss National Science Foundation (no. 31003A-146131) to AHaj and by the Kanton of Zurich.
References
- Haag, A., Gutierrez, P., Buhler, A., Walser, M., Yang, Q., Langouet, M., Kradolfer, D., Frohli, E., Herrmann, C. J., Hajnal, A. and Escobar-Restrepo, J. M. (2014). An in vivo EGF receptor localization screen in C. elegans Identifies the Ezrin homolog ERM-1 as a temporal regulator of signaling. PLoS Genet 10(5): e1004341.
- Phair, R. D., Gorski, S. A. and Misteli, T. (2004). Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Method Enzymol 375: 393-414.
- White, J. and Stelzer, E. (1999). Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol 9(2): 61-65.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Walser, M., Hajnal, A. and Escobar-Restrepo, J. M. (2015). FRAP Analysis of LET-23::GFP in the Vulval Epithelial Cells of Living Caenorhabditis elegans Larvae. Bio-protocol 5(11): e1489. DOI: 10.21769/BioProtoc.1489.
Category
Cell Biology > Cell imaging > Fluorescence
Cell Biology > Cell imaging > Confocal microscopy
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









