- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fluorescence-based CAPS Multiplex Genotyping on Capillary Electrophoresis Systems
(*contributed equally to this work) Published: Vol 5, Iss 10, May 20, 2015 DOI: 10.21769/BioProtoc.1472 Views: 10101
Reviewed by: Fanglian HeKabin XieKanika Gera

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Experimental Pipeline for SNP and SSR Discovery and Genotyping Analysis of Mango (Mangifera indica L.)
Michal Sharabi-Schwager [...] Ron Ophir
Aug 20, 2016 11559 Views

Investigation of Transposon DNA Methylation and Copy Number Variation in Plants Using Southern Hybridisation
Vivek Hari Sundar G. and P. V. Shivaprasad
Jun 5, 2022 3411 Views

CAPS-Based SNP Genotyping for Nitrogen-Response Phenotypes in Maize Hybrids
Jannis Jacobs [...] Peter K. Lundquist
Dec 20, 2025 550 Views
Abstract
Recent advances in next-generation sequencing techniques allow the detection of a large number of SNPs and their use in a high throughput manner. However, Cleaved Amplified Polymorphic Sequences (CAPSs) still play a significant role as complement to other high throughput methods for SNP genotyping. Therefore, new methods focusing on the acceleration of this type of markers are highly desirable. The combination of the classical CAPS technique and a M13-tailed primer multiplexing assay was used to develop an agarose gel free protocol for the analysis of SNPs via restriction enzyme digestion. PCR products were fluorescence labeled with a universal M13 primer and subsequently digested with the appropriate restriction endonuclease. After mixing differently labeled products, they were detected on a capillary electrophoresis system. This method allows the cost-effective genotyping of several SNPs in a multiplexed manner at an overall low cost in a short period of time. Additionally, this method could be efficiently combined with the simultaneous detection of SSRs at the same electrophoresis run resulting in a procedure well suited for marker-based selection procedures, genotyping of mapping populations and the assay of genetic diversity.
Keywords: CAPSMaterials and Reagents
- DNA (25 ng/µl) (e.g. from two-weeks old barley seedlings; or according to PCR practice of each particular organism)
- PCR reagents
- 0.5 U of Taq FIREPol® DNA polymerase (Solis Biodyne FIREPol®, catalog number: 01-01-01000 ; any other suppliers should be also satisfactory) with the corresponding 10x PCR buffer (supplied with FIREPol® DNA polymerase)
- 25 mM MgCl2 (supplied with FIREPol® DNA polymerase)
- dNTPs (10 µM each) (Thermo Fisher Scientific, catalog number: R0182 )
- Forward specific primer tailed at the 5´end with a universal M13 tail (1µM) (5´- CACGACGTTGTAAAACGAC-3´) (desalted) (Microsynth)
- Reverse specific primer (10 µM) (desalted) (Microsynth)
- Fluorescence (6-FAM, HEX, NED or Cy5, D2, D3) labeled primer with a complementary sequence to the M13 tail (10 µM) (Metabion, Planegg/Steinkirchen)
- 0.5 U of Taq FIREPol® DNA polymerase (Solis Biodyne FIREPol®, catalog number: 01-01-01000 ; any other suppliers should be also satisfactory) with the corresponding 10x PCR buffer (supplied with FIREPol® DNA polymerase)
- Restriction analysis
- Restriction endonucleases (New England Biolabs or Fermentas)
- 1x Buffer restriction endonucleases (New England Biolabs or Fermentas)
- Restriction endonucleases (New England Biolabs or Fermentas)
- 1.5% agarose (Sigma-Aldrich, catalog number: A9539 )
- Ethidium bromide (Roche Diagnostics, catalog number: HP46.2 )
- DNA loading buffer 3x [6x; 30% (v/v) glycerol, 0.25% (w/v) bromophenol blue, 0.25% (w/v) xylene cyanol FF]
- Analysis on ABI PRISM 3100 Genetic Analyzer Separation Gel (Applied Biosystems, catalog number: 4363929 )
- ROTISOLV® HPLC gradient grade water (Roche Diagnostics, catalog number: A511.3 )
- HiDiTMFormamide as Sample Loading Buffer (Applied Biosystems, catalog number: 4311320 )
- GeneScanTMROX size standard (Applied Biosystems, catalog number: 401734 )
- 96 well half skirted PCR plates (Kisker, catalog number: G060/H/1E/OA-SS )
- ROTISOLV® HPLC gradient grade water (Roche Diagnostics, catalog number: A511.3 )
- Analysis on Beckman Coulter CEQTM 8000 Genetic Analysis System
- GenomeLabTM Separation Gel LPA I (Beckman Coulter, catalog number: 608010 )
- 96 well half skirted, segmented PCR plates (Kisker, catalog number: G060/H/1E-SG )
- Separation Buffer plates, non-sterile (Beckman Coulter, catalog number: 609844 )
- GenomeLabTM DNA Size Standard 600 (Beckman Coulter, catalog number: 608095 )
- GenomeLabTM Separation Buffer (Beckman Coulter, catalog number: 608012 )
- Formamide as Sample Loading Buffer (Sigma-Aldrich, catalog number: F9037 ) or original Sample Loading Buffer (SLS) (Beckman Coulter, catalog number: 608082)
- GenomeLabTM Separation Gel LPA I (Beckman Coulter, catalog number: 608010 )
- Gel running buffer (see Recipes)
Equipment
- Thermal Cycler (Applied Biosystems)
- Horizontal Electrophoresis system (Bio-Rad Laboratories, Sub-Cell® Model 96 Cell)
- ABI PRISM 3100 Genetic Analyzer or Beckman Coulter CEQTM 8000 Genetic Analysis System
- Heating block, bath or oven for restriction analysis
- PCR 96-well plate or tubes
Software
- GeneMapper® 4.0 Software (Applied Biosystems) or GenomeLabTM GeXP Software (Beckman Coulter)
Procedure
- Prepare a PCR mix in a final volume of 15 µl: 1x PCR buffer, 2.5 mM MgCl2, 0.2 mM dNTPs, 0.02 µM of tailed-forward primer, 0.2 µM of reverse primer, 0.18 µM of fluorescence labeled (6-FAM, HEX, NED or Cy5, D2, D3) M13 primer, 0.5 U of Taq polymerase and 50 ng of DNA.
- Run the following PCR program: 5 min at 94 °C; 12 cycles with 30 sec at 94 °C, 30 sec at 62 °C (touchdown of 0.5 °C / cycle for initial 12 cycles - final annealing of 56 °C for remaining 35 cycles), 30 sec at 72 °C; and a final extension step of 10 min at 72 °C.
- Check 3 µl of the PCR product in an agarose gel (1.5%) stained with ethidium bromide.
- Restriction assay. The presence of restriction sites should be previously assessed by comparing the sequences of target samples. The search for the suitable restriction endonuclease might be carried out with the tool SNP2CAPS (http://pgrc.ipk-gatersleben.de/snp2caps/) (Thiel et al., 2004) or using the web tool NEB cutter (http://nc2.neb.com/NEBcutter2/). The prediction of restriction analysis on each fragment with the corresponding enzyme might be performed at the TAIR website (http://www.arabidopsis.org/). Fragments larger than 600 bp should be discarded.
- Place 12 µl (remaining PCR mix) of the PCR product into a PCR tube.
- Add 1 U of the respective restriction enzyme, together with the specific buffer.
- Incubate in a heat block, bath or oven at the adequate temperature, according to the instructions of enzyme suppliers, for 3 h.
- Place 12 µl (remaining PCR mix) of the PCR product into a PCR tube.
- Fragment analysis on ABI PRISM 3100 Genetic Analyzer or Beckman Coulter CEQTM8000 Genetic Analysis System.
a.Mix equal amounts of each restricted product (6-FAM, HEX, NED or Cy5, D2, D3) for multiplexing. M13-tailed products should be usualy 1/20 diluted with HPLC gradient grade water (ROTISOLV®) in a volume of 20 µl.
b.Make mastermix by mixing HiDi-Formamide and the Rox-Standard for each sample – 14 µl HiDi and 0.04 µl Rox-Standard.
c.Add 14 µl of mastermixes in each well ABI or Beckman PCR-plate (P/N N8010560) and 1 µl of diluted PCR-product.
d.Denaturate the DNA plate for 5 min at 96 °C, and after change of buffer/water in tanks of ABI PRISM Analyzer or Beckman Coulter Analysis System, place into appliance.
- Data analysis with the corresponding software (GeneMapper® 4.0 Software or GenomeLabTM GeXP Software). Only those fragments which retain the fluorescent label will be detected (Figure 1).
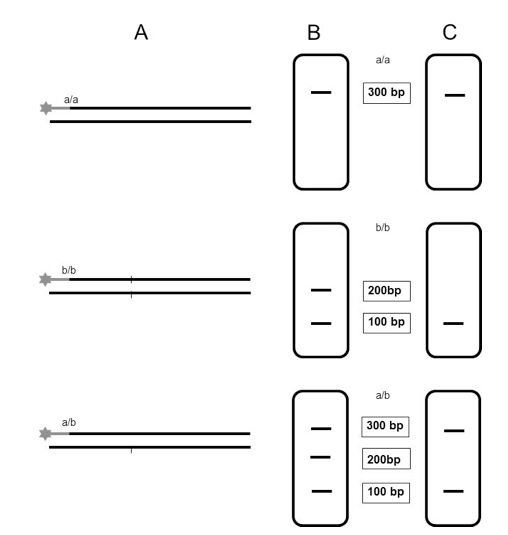
Figure 1. Schematic illustration of the expected results after analysis on capillary electrophoresis systems using the CAPS fluorescence-based multiplexing protocol. A. Digestion of labeled PCR products from two homozygous cultivars (a/a) and (b/b) and from the corresponding heterozygous F1 generation (a/b). B. Separation of digested products on agarose gel. C. Separation of digested products on capillary electrophoresis systems; only those fragments which are detected on the genetic analyzer are represented.
Notes
- This protocol could be efficiently combined with the simultaneous detection of simple sequence repeat (SSR) markers in the same capillary electrophoresis run (Perovic et al., 2013).
Recipes
- Gel running buffer
1x Tris-Borate-EDTA
89 mM Tris base
89 mM Boric acid
2 mM EDTA (pH 8.0)
Acknowledgments
This protocol is adapted from Perovic et al. (2013).
References
- Perovic, J., Silvar, C., Koenig, J., Stein, N., Perovic, D. and Ordon, F. (2013). A versatile fluorescence-based multiplexing assay for CAPS genotyping on capillary electrophoresis systems. Mol Breed 32(1): 61-69.
- Thiel, T., Kota, R., Grosse, I., Stein, N. and Graner, A. (2004). SNP2CAPS: a SNP and INDEL analysis tool for CAPS marker development. Nucleic Acids Res 32(1): e5.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Perovic, J., Silvar, C., Perovic, D., Stein, N. and Ordon, F. (2015). Fluorescence-based CAPS Multiplex Genotyping on Capillary Electrophoresis Systems. Bio-protocol 5(10): e1472. DOI: 10.21769/BioProtoc.1472.
Category
Plant Science > Plant molecular biology > DNA > Genotyping
Molecular Biology > DNA > Genotyping
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









