- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Microscopic Observation, Three-dimensional Reconstruction, and Volume Measurements of Sperm Nuclei
Published: Vol 5, Iss 7, Apr 5, 2015 DOI: 10.21769/BioProtoc.1437 Views: 9443
Reviewed by: Tie LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1691 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views

A Simple Protocol for Periodic Live Cell Observation of Flagellate Stages in the Lichen Alga Trebouxia
Enrico Boccato [...] Mauro Tretiach
Jan 20, 2026 189 Views
Abstract
Karyogamy, a migration of the sperm nucleus toward the egg nucleus and their subsequent nuclear fusion, is an important biological event for initiating zygote formation toward early embryogenesis in angiosperms. However, how the male nucleus approaches and fuses with the female nucleus still remains unclear. Recently, time-lapse measurement of nuclear volume during karyogamic events revealed that the sperm nucleus enlarges during contact with the egg nucleus via possible one-directional migration of egg chromatin into sperm nucleus (Ohnishi et al., 2014). Here, we describe the protocol for microscopic observation, three-dimensional reconstruction, and volume measurements of sperm nuclei in rice zygotes/fused gametes, which are produced by an in vitro fertilization system (Uchiumi et al., 2006; Uchiumi et al., 2007). The present protocol will be applied for monitoring nuclear dynamics in cells during cell division, differentiation, de-differentiation and polarity formation as well as karyogamy progression.
Keywords: Egg cellMaterials and Reagents
- Transgenic rice plants (Oryza sativa L. cv. Nipponbare) expressing SUN2-GFP or H2B-RFP fusion proteins that are targeted to nuclear membrane or nuclear chromatin, respectively (Ohnishi et al., 2014) (see Note 1)
- Mannitol (Wako Chemicals USA, catalog number: 133-00845 )
- 370 mosmol/kg H2O (330 mM) mannitol solution (autoclaved)
- 450 mosmol/kg H2O (385 mM) mannitol solution (autoclaved)
- Mineral oil (Sigma-Aldrich, catalog number: M8410-100ML )
Equipment
- Inverted fluorescent microscope (OLYMPUS, model: BX-71 )
- Confocal laser scanning (CLS) microscope (ZEISS, model: LMS 710 )
- Non-treated plastic dishes with diameter of 3.5 cm (Iwaki, catalog number: 1000-035 )
- Glass base dishes with diameter of 3.5 cm (Iwaki, catalog number: 3971-035 ) (see Note 2)
- Coverslips (24 x 40 mm) (Thermo Fisher Scientific, catalog number: 125485J ) (siliconized at the edges with 5% dichloromethylsilane in 1,1,1-trichloroethane) (see Note 3)
- Glass capillaries made from 50 μl aspirator tubes (Drummond Scientific Company, catalog number: 2-000-050 ) (Figure 1A-B) (see Note 4)
- Manual handling injector (ST Science, type UJB)
- Environmental chamber (Koito Industries Ltd., catalog number: K30-7248 )
Software
- IMARIS software (Bitplane)
Note: A core scientific software module that delivers the necessary functionality for data management, visualization, analysis, segmentation and interpretation of 3D and 4D microscopy datasets.
Procedure
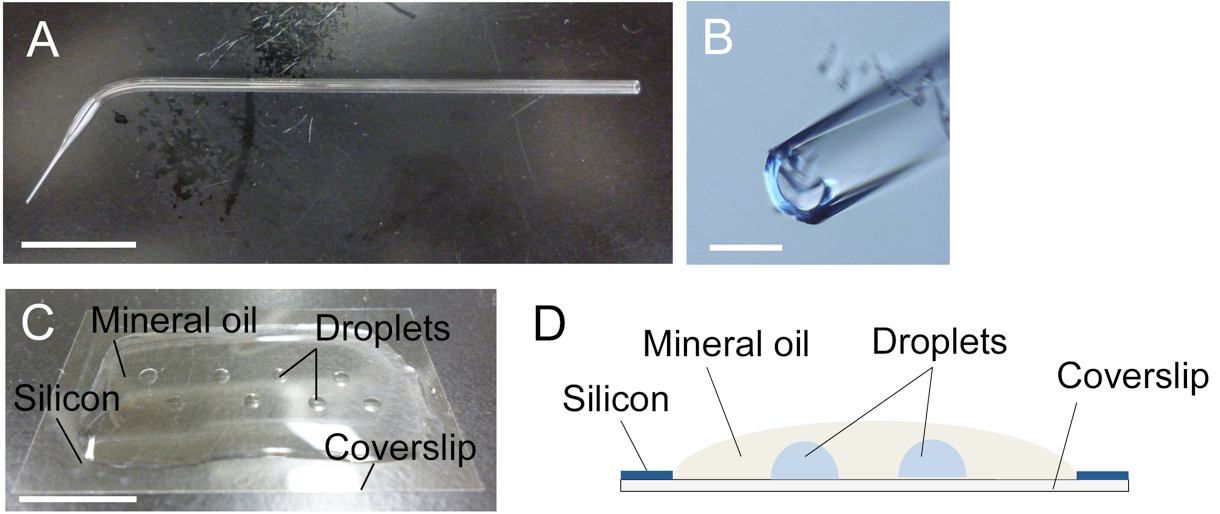
Figure 1. Glass capillary (A, B) and mannitol droplets on coverslip (C, D). A. A glass capillary drawn by hand. Tip region was boxed. B. Tip opening of the glass capillary in panel A. C and D. A photo (C) and an image (D) of mannitol droplets inside the mineral oil over the siliconized coverslip. Bars = 1 cm in A and C, 200 μm in B.
- Sperm cell isolation and microscopic observation of sperm nuclear membrane
- Overlay the siliconized coverslip with 0.3 ml mineral oil and make several 1-2 μl mannitol droplets (370 mosmol/kg H2O) inside the mineral oil on the coverslip (Figure 1C) using a glass capillary connected to a handling injector. Take care that the droplets have no access to the air (Figure 1D).
- Dissect an unopened flower from the transgenic rice plants expressing SUN2-GFP to obtain anthers under a dissecting microscope (Figure 2A and B).
- Transfer anthers into a 3.5 cm plastic dishe filled with 3 ml of mannitol solution (370 mosmol/kg H2O). Break anthers in mannitol solution with forceps to free pollen grains (Figure 2C).
- Pollen grains burst due to the osmotic shock and release sperm cells. Identify the sperm cells under an inverted microscope (Figure 3A). By using a glass capillary connected to a handling injector, collect sperm cells and transfer into mannitol droplets prepared in step A1.
- SUN2-GFP localizing in nuclear membrane is visualized with an inverted fluorescence microscope with 460-490 nm excitation and 510-550 nm emission wavelengths (Figure 3B and C).
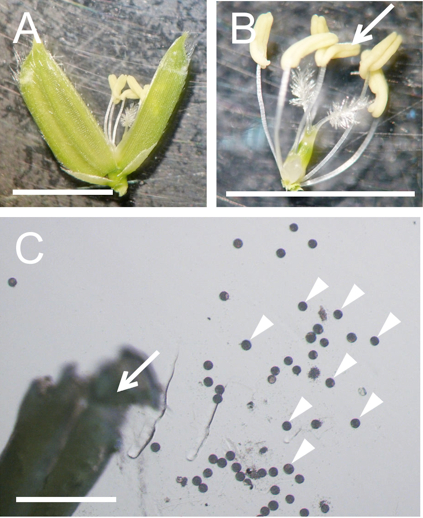
Figure 2. Preparation of pollen grains from rice flower. A. A mature flower whose lemma and palea were opened. B. A pistil connected with six stamens isolated from the flower in panel A. C. A broken anther and released pollen grains in mannitol solution. Arrows in panels B and C and arrowheads in panels C indicate anthers and pollen grains, respectively. Bars = 5 mm in A and B, 500 µm in C.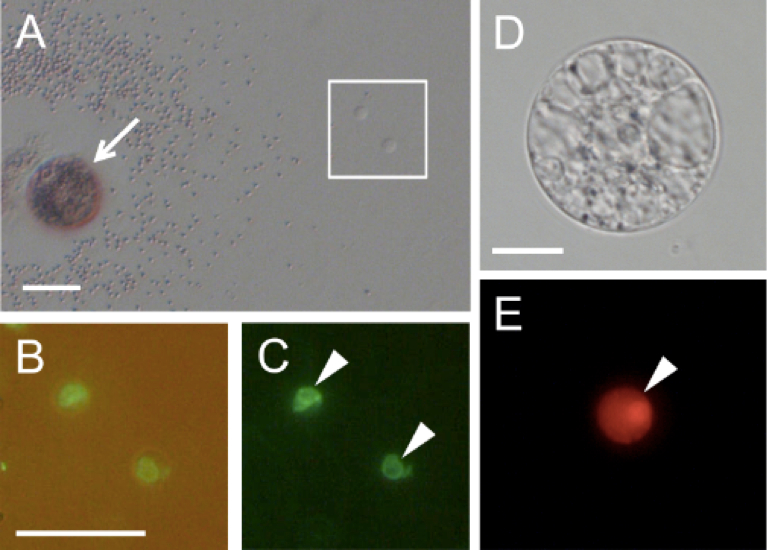
Figure 3. Rice gametes expressing the SUN2-GFP or histone H2B-GFP fusion protein. A. A pollen grain expressing SUN2-GFP released its content in mannitol solution. B and C. Two sperm cells enclosed within the square in panel A were visualized under fluorescence microscopy. Panel C contains the merged bright-field and fluorescent images. D and E. An egg cell isolated from transgenic rice expressing histone H2B-RFP was visualized under bright-field (D) and fluorescence (E) microscopy. Arrow indicates a pollen grain releasing its content. Arrowheads in panels C and E indicate sperm nucleus and egg nucleus, respectively. Bars = 20 μm
- Overlay the siliconized coverslip with 0.3 ml mineral oil and make several 1-2 μl mannitol droplets (370 mosmol/kg H2O) inside the mineral oil on the coverslip (Figure 1C) using a glass capillary connected to a handling injector. Take care that the droplets have no access to the air (Figure 1D).
- In vitro fertilization with rice gametes and LSM observation of gamete nuclear membrane and nucleus in the fused gametes/zygotes
- Overlay the glass base dish with 0.2 ml mineral oil, and make a 1-2 μl mannitol droplet (450 mosmol/kg H2O) using a glass capillary connected with handling injector.
- Isolate egg cells from transgenic rice plants expressing H2B-RFP according to the previous protocol (Okamoto, 2011). By using a glass capillary connected to a handling injector, transfer an egg cell in a mannitol droplet on a coverslip prepared in step A1 (Figure 3D and E).
- Collect sperm cells expressing SUN2-GFP as described in step A, and transfer a sperm cell into a mannitol droplet with an egg cell from step B2.
- Electro-fuse the sperm cell and the egg cell in the droplet according the previous protocol (Okamoto, 2011). Briefly, an egg cell was fixed at one electrode under an alternating current field, and a sperm cell was aligned with the egg cell. Cell fusion is induced by applying a single negative direct current pulse, resulting in production of a fused gamete (zygote).
- Transfer the resultant zygote to a mannitol droplet on the glass base dish prepared in step B1. Observe the zygote under the LMS 710 CLS microscope using an oil immersion objective lens with 488-nm excitation and 505-530-nm emission wavelengths for SUN2-GFP and with 543-nm excitation and >560-nm emission wavelengths for H2B-RFP.
- Obtaining Z-stack serial images of sperm nuclei by LSM observations are described below.
Image size: 1,024 pixel (X-axis) X 1,024 pixel (Y-axis), Digital zoom: 3.8 fold, Pinhole: 90 μm, Laser power (488-nm): 0.3%, Laser power (543-nm): 0.3%, Number of serial image (Z-axis): 12-16, Z-stack thickness: 0.6 μm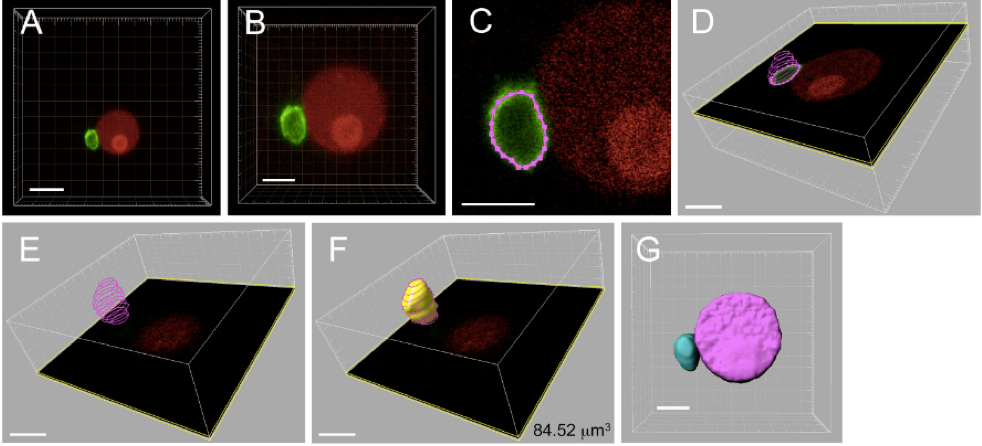
Figure 4. Three-dimensional reconstruction and volume measurement of sperm nucleus within a zygote. The zygote produced by electro-fusion of a sperm cell expressing SUN2-GFP and an egg cell expressing H2B-RFP was observed under a CLS microscope. A. The 3D image automatically constructed by the IMARIS software using Z-stack images from CLS microscope observation. B. Magnified image of panel A. C. Outline of sperm nucleus, represented in pink colored dotted line, was marked by spotting points on the SUN2-GFP labeled nuclear membrane for each Z-stack images. D. Merged outlines of a sperm nucleus from six Z-stack images. E. Merged outlines for a sperm nucleus composed of 12 Z-stack images. F. The 3D image constructed from the merged outlines of a sperm nucleus. The number in the panel represents the volume of sperm nucleus. G. The 3D image of sperm and egg nuclei. Bars = 10 μm in A, 5 μm in B-G
- Overlay the glass base dish with 0.2 ml mineral oil, and make a 1-2 μl mannitol droplet (450 mosmol/kg H2O) using a glass capillary connected with handling injector.
- Three-dimensional reconstruction and volume measurements of sperm nucleus during karyogamy
- Open Z-stack serial images of sperm nuclear membrane using IMARIS software. A 3D image is automatically generated (Figure 4A).
- Adjust the size of the 3D image for image manipulation (software operation: Edit-Crop 3D) (Figure 4B).
- Filter out noise signal from the 3D image created in step C2 (software operation: Image processing-Image smoothing).
- In the manual 3D construction mode (software operation: Surpass-Surface-Manual), generate an outline of sperm nucleus by spotting points on the SUN2-GFP labeled nuclear membrane for each Z-stack image (software operation: Draw-Contour-Mode- Click) (Figure 4C-E).
- Reconstruct the 3D image using the manually outlined sperm nucleus from step C4 (software operation: Create surface) (Figure 4E-F), and measure the volume of sperm cell nucleus (software operation: Imaris Maesurement Pro-Volume) (Figure 4F-G).
- Open Z-stack serial images of sperm nuclear membrane using IMARIS software. A 3D image is automatically generated (Figure 4A).
Notes
- Rice plants are grown in environmental chamber at 26 °C in a 13/11 h light/dark cycle with a photosynthetic photon flux density of 150-300 μmol photons/m2/s.
- Glass of the glass base dishes should be non-coated, as using coated glass base will result in attachment of the cells to the surface of the glass.
- Coverslips should be also non-coated, as using coated coverslips will result in attachment of the cells to the surface of the coverslip.
- Tip openings were drawn by hand to 150-250 μm.
Acknowledgments
This work was supported, in part, by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Nos. 21112007 and 26113715 to T.O.) and from the Japan Society for the Promotion of Science (No. 25650083 to T.O.). We thank Ms. Hiroki Maeda and Ms. Tomoko Mochizuki (Tokyo Metropolitan University) for technical assistance, and Dr. Min Yvonne Kim (University of California, Berkeley) for critical reading of the protocol.
References
- Ohnishi, Y., Hoshino, R. and Okamoto, T. (2014). Dynamics of male and female chromatin during karyogamy in rice zygotes. Plant Physiol 165(4): 1533-1543.
- Okamoto T. (2011). In vitro fertilization with isolated rice gametes: production of zygotes and zygote and embryo culture. Methods Mol Biol 710: 17-27.
- Uchiumi, T., Komatsu, S., Koshiba, T. and Okamoto, T. (2006). Isolation of gametes and central cells from Oryza sativa L. Sex Plant Reprod 19(1): 37-45.
- Uchiumi, T., Uemura, I. and Okamoto, T. (2007). Establishment of an in vitro fertilization system in rice (Oryza sativa L.). Planta 226(3): 581-589.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ohnishi, Y. and Okamoto, T. (2015). Microscopic Observation, Three-dimensional Reconstruction, and Volume Measurements of Sperm Nuclei. Bio-protocol 5(7): e1437. DOI: 10.21769/BioProtoc.1437.
- Ohnishi, Y., Hoshino, R. and Okamoto, T. (2014). Dynamics of male and female chromatin during karyogamy in rice zygotes. Plant Physiol 165(4): 1533-1543.
Category
Plant Science > Plant cell biology > Cell imaging
Plant Science > Plant cell biology > Tissue analysis
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









