- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of the Secondary Structure of an RNA fragment in Solution: Selective 2`-Hydroxyl Acylation Analyzed by Primer Extension Assay (SHAPE)
Published: Vol 5, Iss 2, Jan 20, 2015 DOI: 10.21769/BioProtoc.1386 Views: 14418
Reviewed by: Tie LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

General Maintenance and Reactivation of iSLK Cell Lines
Ariana C. Calderón-Zavala [...] Ekaterina E. Heldwein
Jun 5, 2025 1896 Views

Inducible HIV-1 Reservoir Reduction Assay (HIVRRA), a Fast and Sensitive Assay to Test Cytotoxicity and Potency of Cure Strategies to Reduce the Replication-Competent HIV-1 Reservoir in Ex Vivo PBMCs
Jade Jansen [...] Neeltje A. Kootstra
Jul 20, 2025 2460 Views

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3038 Views
Abstract
This protocol describes the methodology for the determination of the secondary structure of an RNA fragment in solution using Selective 2´-Hydroxyl Acylation analyzed by Primer Extension, abbreviation SHAPE. It consists in the very fast chemical modification of flexible and therefore possibly single-stranded nucleotides in a sequence-independent manner using benzoyl cyanide (BzCN), forming 2´-O-adducts. The modifications in the RNA are then analyzed by primer extension. Reverse transcriptase is blocked by the 2´-O-adducts formed. The advantage of the method is, first, that not each RNA molecule studied but the primer used in the extension reaction is labelled and, second, that the resulting cDNA analyzed in sequencing gels is much more stable than the modified RNA.
Keywords: RNA secondary structureMaterials and Reagents
- Yeast tRNA (Life Technologies, Ambion®, catalog number: AM7119 )
- MEGAshortscript T7 Transcription Kit (Life Technologies, Ambion®, catalog number: AM1354 )
- Oligonucleotide (50 μM)
- Recombinant RNase inhibitor (Takara Bio Company, catalog number: 2313A )
- Potassium chloride (Sigma-Aldrich, catalog number: P9541 )
- HEPES (Sigma-Aldrich, catalog number: H4034 )
- Magnesium chloride (Sigma-Aldrich, catalog number: 63063 )
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418 )
- Benzoyl cyanide (Sigma-Aldrich, catalog number: 115959 )
Note: Keep in desiccator! - 3 M sodium acetate (pH 5.5) (Life Technologies, Ambion®, catalog number: AM9740 )
- T4 Polynucleotide kinase (New England Biolabs, catalog number: M0201S )
- ATP ([γ-32P]- 6,000 Ci/mmol 10 mCi/ml Lead, 250 µCi) (PerkinElmer, catalog number: NEG002Z250UC )
- Micro Bio-Spin P-30 Gel Columns Tris Buffer (Bio-Rad Laboratories, catalog number: 732-6250 )
- dNTPs mixture for primer extension (2.5 mM each) (Takara Bio Company, catalog number: 4030 )
- ddNTPs (set 5 mM) (GE Healthcare, catalog number: 27-2045-01 )
- dNTPs (set 100 mM diluted to 10 mM for ladder) (Life Technologies, catalog number 10297-018 )
- SuperScript II (Life Technologies, InvitrogenTM, catalog number: 18064-014 )
- Gel loading buffer II (Life Technologies, Ambion®, catalog number: AM8546G )
- Sodium hydroxide (Sigma-Aldrich, catalog number: 221465 )
- Ammonium persulfate (APS) (Bio-Rad Laboratories, catalog number: 161-0700 )
- 100% ethanol
- Biospin columns (Bio-Rad Laboratories, catalog number: 732-6250)
- 10x TBE buffer (see Recipes)
- Denaturing polyacrylamide gel electrophoresis (PAGE) (see Recipes)
- Sequencing ladders (ddNTP/dNTP mix) (see Recipes)
- 5x SHAPE buffer (see Recipes)
- 0.4 M benzoyl cyanide (see Recipes)
Equipment
- Standard laboratory equipment
- Nanodrop device
- Incubator or water bath
- Sequencing gel electrophoresis system
- Power supply
- Vacuum pump
- Gel dryer (Bio-Rad Laboratories, model: 583 )
- Phosphorimaging instrument and screen
- Typhoon 9410 scanner
- The UreaGel System (National Diagnostics, catalog number: EC-833 )
Software
- Image analysis software (Bio-Rad Laboratories, Quantity One; SAFA footprinting software, https://simtk.org/home/safa)
- RNA secondary structure prediction
Procedure
- Cloning of RNA to be analyzed in SHAPE cassette
RNA segment was cloned into SHAPE cassette (Figure 1) (Wang et al., 2010). SHAPE cassette has been checked to ensure that it is not prone to forming stable base pairing interactions with the internal sequence.
We have design a structure cassette that contains flanking sequences that allow evaluate all positions within the RNA of interest. The primer binding site of this cassette efficiently binds to a cDNA primer.
Figure 1. Schematic representation of the construct used for the determination of secondary RNA structure by SHAPE technology. The sequence fragment of the RNA to be analyzed is cloned between the EcoRI and HpaI restriction sites of the SHAPE cassette (Wang et al., 2010). - Preparation of RNA by in vitro transcription
- Linearized DNA plasmid (≈0.5 µg) with SmaI used as template for in vitro transcription with MEGAshortscript (transcription protocol in user guide).
- After transcription, purification was made by phenol:chloroform extraction and alcohol precipitation as described in protocol. Alcohol precipitation was carried out overnight at - 20 °C with 3 M sodium acetate (pH 5.2) instead ammonium acetate.
- RNA recovery by centrifugation, followed by wash with 70% ethanol and dissolve pellet in 20 µl nuclease-free water.
- Linearized DNA plasmid (≈0.5 µg) with SmaI used as template for in vitro transcription with MEGAshortscript (transcription protocol in user guide).
- SHAPE analysis
- RNA refolding
Need 500 ng to 1 µg RNA per reaction. - Resuspend RNA in water. For example for 4 reactions resuspend RNA to make 40 µl total volume in an Eppendorf tube.
- Denaturation of RNA, 94 °C/1 min.
- Incubate on ice, 2 min.
- Divide 10 µl of refolded RNA into each tube.
- Add 30 µl of nuclease free water supplemented with 0.5 µl RNAse inhibitor into each tube.
- Add 10 µl of 5x SHAPE buffer and incubate, 30 °C/30 min.
- Modification
Add 5.5 µl of DMSO (untreated) or BzCN (treated), 2 min. - Add 6 µl of 3 M NaOAc (pH 5.2) in each epp.
- Add 2 µl of tRNA yeast at 1.9 mg/ml + 190 µl of 100% EtOH, -20 °C, overnight.
Next day - Centrifuge 14,000 rpm 4 °C, 40 min.
- Wash using 1ml of 70% ethanol and centrifuge 14,000 rpm, 4 °C, 10 min.
- Dry RNA pellet and resuspend in 20 µl. Quantify in Nanodrop. Continue with primer extension only with RNAs ≥ 100 ng/µl, 15 min.
- Label primer37 °C, 30 min
dH2O 33 µl Primer (150 pmol) 3 µl 10x PNK buffer 6 µl γP32 ATP 15 µl PNK 3 µl - Purify using biospin columns.
- Measure counts in a liquid scintillation counter if 107 cpm activity proceed to probing.
- Primer extension
RNA modified Sequencing ladder RNA 500 ng RNA no treated 200 ng dNTPs (2.5 mM) 2 µl dNTPs/ddNTPs 2 µl Labeled primer 2 µl Labeled primer 2 µl dH2O Total 12 µl Total 12 µl - Incubate on ice, 2 min.
- Incubate at room temperature, 5 min.
- Cocktail RT (reverse transcription)42 °C, 30 min
0.1 mM DTT 2 µl RT buffer 4 µl RNase inhibitor 0.2 µl RT 0.1 µl dH2O 2 µl - Add 4 µl of 1 M NaOH, 95 °C, 30 min.
- Add of Gel loading buffer II (see Item 18 in Materials and Reagents) (15-20 µl), 95 °C, 4 min.
- Mix, spin and incubate on ice before loading onto the gel, 3 min.
- Pre-run gel: Load 2 µl Gel loading buffer II and conduct electrophoresis at constant power of 60 W, 15 min.
- Load samples in gel: 2 µl/lane. Conduct electrophoresis at constant power of 60 W.
- Run gel (you may do a long run approximately 3 h for resolving 5´end or a short run approx. 2 h for resolving 3’ end or your molecule), 2-3 h.
- Remove one of the glasses plates, allowing the gel to remain attached to the second plate. Press a sheet of Whatman paper on top of the gel. The gel will adhere to the Whatman paper and can be peeled away from the remaining glass plate. Cover the gel with plastic wrap.
- Dry the gel, 2 h.
- Expose to phosphorimager screen, ≈ 15 h.
- Scan in Typhoon 9410 scanner at 50 micron resolution (Figure 2), 10 min.
- Use SAFA Footprinting Software for analyze bands: https://simtk.org/docman/view.php/69/496/SAFAUserGuide_v1_1.pdf.
The protocols implemented in SAFA have five steps: (a) lane identification, (b) gel rectification, (c) band assignment, (d) model fitting and (e) band-intensity normalization.
Software will generate normalized bands intensities values (reactivity data). - Use reactivity data from SAFA and classify it in “High, medium and low reactivity” according with normalized band intensity value from each nucleotide. Use “x” or “X” to indicate reactive nucleotides and “.” to indicate unreactive. Fill it in point 4 (Single-Stranded Chemical/Enzymatic Reactivity Data) of MC-Fold http://www.major.iric.ca/MC-Fold/ (Figure 3).
- MC-Fold will generate the top 20 structures.
- Choose structures that they are more fit of your reactivity data.
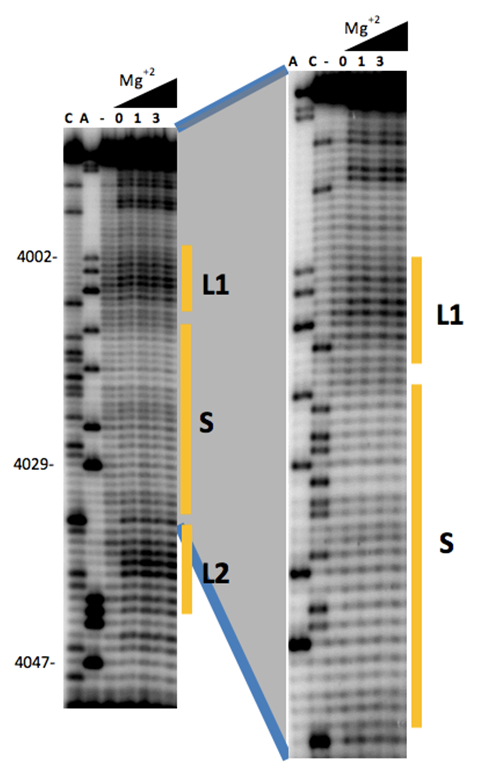
Figure 2. Secondary structure probing of a cap-independent translation enhancer (3´-CITE). Structure probing by SHAPE of the first 65 nt of the 3´-UTR of MNSV-N, including the new 3´-CITE. Primer extension products separated on denaturing PAGE of RNA treated (lanes 4-6) or untreated (lane 3) with BzCN. Concentrations of Mg2+ (mM) are indicated above lines 4-6. The sequencing ladder was generated by reverse transcription of unmodified RNA in the presence of dideoxyCTP (ddCTP) or ddATP. Positions of nucleotides A4002, A4029 and A4047 are indicated on the left.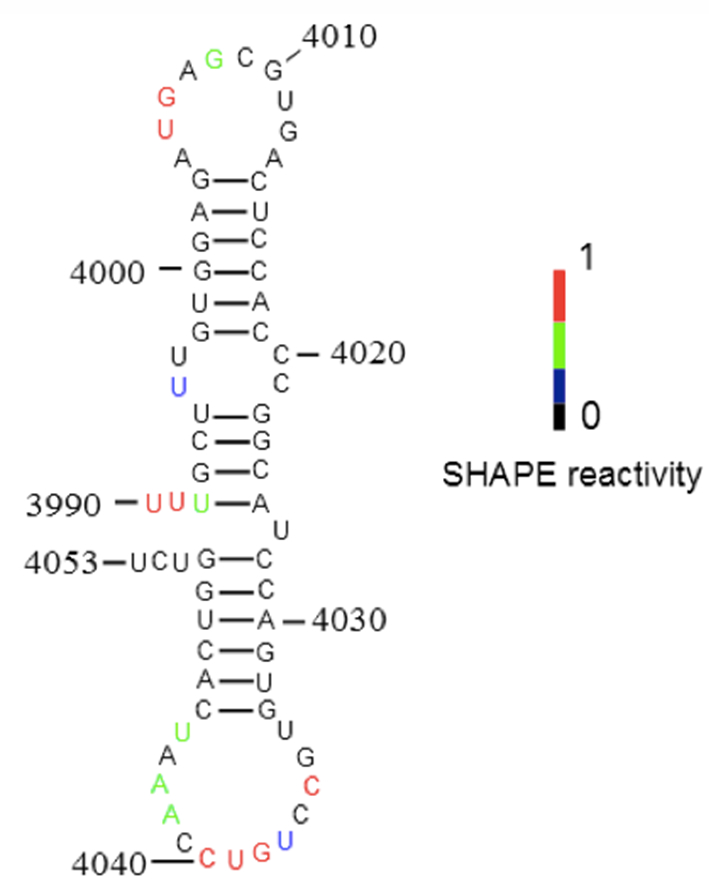
Figure 3. Secondary structure of the new 3´-CITE probed in Figure 2. SHAPE reactivity of nucleotides superimposed on secondary structure predicted by Mfold. Color-coded bases indicate the levels of BzCN modification, with warmer color indicating greater modification (inset).
- RNA refolding
Recipes
- 10 x TBE buffer
108 g of Tris base
55 g of boric acid
40 ml of 0.5 M of EDTA (pH 8.0) - Denaturing polyacrylamide gel electrophoresis (PAGE)
Gel sequencing system
8 M urea
8-15% 37% acrylamide-bisacrylamide 29:1
1x TBE
10% ammonium persulfate (APS)
N, N, N´, N´-tetramethylethylenediamine (TEMED)
Note: Alternatively, The UreaGel System can be used to prepare gels of varying percentage. - Sequencing ladders (ddNTP/dNTP mix)
Note: If signal is too strong add less ddNTP and vice versa.Sequencing ladders A (ul) T (ul) C (ul) G (ul) dATP (10 mM) 1 2 2 2 dTTP 2 0.5 2 2 dCTP 2 2 1 2 dGTP 2 2 2 1 ddATP (5 mM) 20 ddTTP 20 ddCTP 10 ddGTP 20 dH2O 13 13.5 23 13 - 5x SHAPE buffer
Note: The buffer should be optimized for each individual RNA to be studied.1x 5x KCl 100 mM 20 ml 1 M KCl HEPES KOH (pH 7.5) 50 mM 10 ml 1 M HEPES KOH (pH 7.5) MgCl2 8 mM 1.6 ml 1 M MgCl2 8.4 ml H2O - 0.4 M benzoyl cyanide
Weight 0.0524 g and dissolve in 1 ml of DMSO
Acknowledgments
This work was supported by grants AGL2009-07552/AGR from Ministerio de Ciencia e Innovación (Spain) and EUI2009-04009 of the transnational (Germany, France, Spain and Portugal) cooperation within the 2009 PLANT-KBBE initiative with funding from Ministerio de Ciencia e Innovación (Spain). Manuel Miras was recipient of a predoctoral fellowship from Ministerio de Ciencia e Innovación (Spain). JJK was funded by grant 2011-67012-30715 from the USDA National Research Initiative.
This protocol was adapted from previous work (Wilkinson et al., 2006; Mortimer and Weeks, 2007).
References
- Kraft, J. J., Treder, K., Peterson, M. S. and Miller, W. A. (2013). Cation-dependent folding of 3' cap-independent translation elements facilitates interaction of a 17-nucleotide conserved sequence with eIF4G. Nucleic Acids Res 41(5): 3398-3413.
- Miras, M., Sempere, R. N., Kraft, J. J., Miller, W. A., Aranda, M. A. and Truniger, V. (2014). Interfamilial recombination between viruses led to acquisition of a novel translation‐enhancing RNA element that allows resistance breaking. New Phytol 202(1): 233-246.
- Mortimer, S. A. and Weeks, K. M. (2007). A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc 129(14): 4144-4145.
- Wang, Z., Kraft, J. J., Hui, A. Y. and Miller, W. A. (2010). Structural plasticity of Barley yellow dwarf virus-like cap-independent translation elements in four genera of plant viral RNAs. Virology 402(1): 177-186.
- Wang, Z., Parisien, M., Scheets, K. and Miller, W. A. (2011). The cap-binding translation initiation factor, eIF4E, binds a pseudoknot in a viral cap-independent translation element. Structure 19(6): 868-880.
- Wilkinson, K. A., Merino, E. J. and Weeks, K. M. (2006). Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1(3): 1610-1616.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Miras, M., Sempere, R. N., Kraft, J. J., Miller, W. A., Aranda, M. A. and Truniger, V. (2015). Determination of the Secondary Structure of an RNA fragment in Solution: Selective 2`-Hydroxyl Acylation Analyzed by Primer Extension Assay (SHAPE). Bio-protocol 5(2): e1386. DOI: 10.21769/BioProtoc.1386.
Category
Microbiology > Microbe-host interactions > Virus
Plant Science > Plant molecular biology > RNA > RNA structure
Molecular Biology > RNA > RNA structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









