- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Imaging and Measurement of Nanomechanical Properties within Primary Xylem Cell Walls of Broadleaves
Published: Vol 4, Iss 24, Dec 20, 2014 DOI: 10.21769/BioProtoc.1360 Views: 13232
Reviewed by: Arsalan DaudiMasahiro MoritaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2322 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views
Abstract
A technique of atomic force microscopy (AFM) called PeakForce quantitative nanomechanical mapping (PeakForce QNM) is an efficient tool for the quantitative mechanobiological imaging of fibrillar aggregate, human epidermal cell and woody plant cell wall topography (Sweers et al., 2011; Heu et al., 2012; Ďurkovič et al., 2012; Ďurkovič et al., 2013). Here, we describe a detailed protocol for the measurement of nanomechanical properties of primary xylem cell walls in woody plants, for the determination of reduced Young’s modulus of elasticity (MOE), adhesion, deformation, and energy dissipation (Figure 1). This new technique provides direct control of the maximum loading force and the deformation depth in cell wall samples keeping indentations small, while at the same time eliminating damaging lateral forces in order to preserve both the AFM tip and plant sample. High-resolution and non-destructive imaging shed new quantitative mechanistic insights into the structural biology of woody plant cell walls. This procedure can also be adapted for other biological samples with a varying range of stiffness.
Keywords: Atomic force microscopy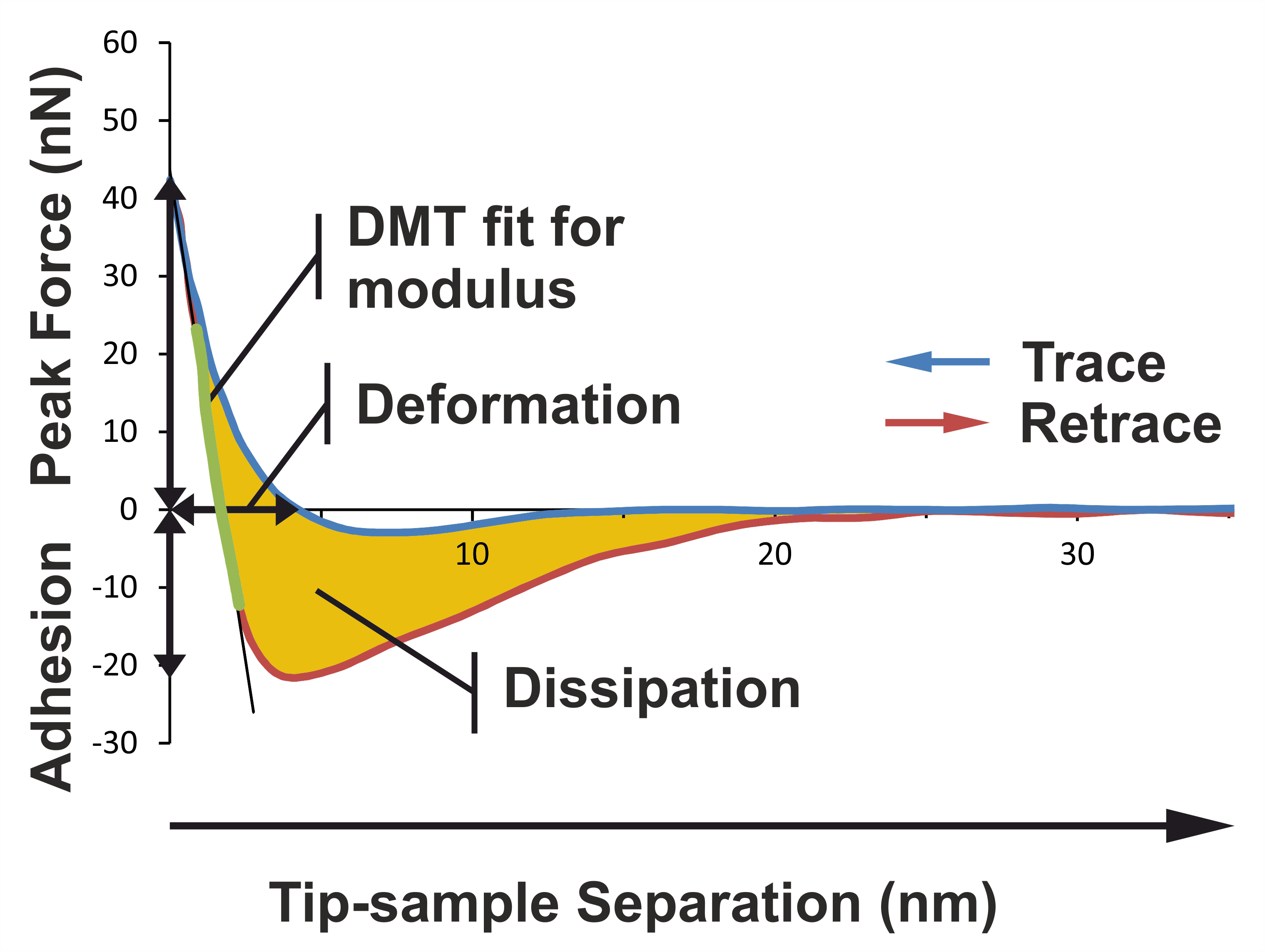
Figure 1. Basic principles illustrating how the different nanomechanical properties are extracted from the force curve. The image shows a typical force curve for a primary xylem cell wall sample in the Dutch elm disease-sensitive hybrid 'Groeneveld'. PeakForce QNM mode performs a very fast force curve at every pixel in the image. Analysis of force curve data is done on the fly, providing a map of multiple nanomechanical properties that has the same resolution as the height image. The adhesion is the vertical distance between the base line and the lowest portion of the retraction curve. The deformation is the horizontal distance between the contact point and the turn-away point (representing maximum indentation). The Young’s modulus of elasticity can be extracted by extrapolating the linear portion of the retraction curve after the contact point using a Derjaguin, Muller, Toporov (DMT) model fit. The slope of the linear portion is determined according to the least square method procedure. The energy dissipation can be calculated by integrating the area between the two curves.
Materials and Reagents
- Leaf midribs from broadleaves (Ulmus spp., Sorbus spp.)
- Glutaraldehyde solution, 25% in H2O (Grade II) (Sigma-Aldrich, catalog number: G6257 )
- Ethanol solutions (Analytical Reagents)
Note: 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, each of them prepared by diluting of 96% ethanol in a 0.1 M cacodylate buffer, and 100% ethanol.
- 25% glutaraldehyde (Grade I) (Sigma-Aldrich, catalog number: G5882 )
- Xylene (Analytical Reagents) or alternative cleaners with a lower toxicity (for example D-limonene)
Note: Work with xylene or D-limonene only in a properly operating and certified fume cupboard.
- Paraplast plus (Sigma-Aldrich, catalog number: P3683 )
- (3-Aminopropyl) triethoxy-silane (Sigma-Aldrich, catalog number: A3648 )
- 0.1 M cacodylate buffer (see Recipes)
- Circle glass slides coated with (3-aminopropyl)triethoxy-silane (see Recipes)
- 1x phosphate buffered saline (PBS) (see Recipes)
- Glutaraldehyde fixative solution (see Recipes)
Equipment
- Razor blades (fine tweezers and heated forceps)
- pH meter
- Exicator with a vacuum pump
- Petri dishes
- Laboratory drying chamber
- Retracting base sledge microtome (Series 8000) (Bright Instrument Co.)
- Circle glass slides (11 mm in diameter, 1 mm thick)
- Slide warmer
- Steel AFM sample mounting disks
- Sample adhesive pads
- Silicon probes MPP-12120 (model: TAP150A ) having a 180° rotated tip and the spring constant at least 5 N/m (Bruker AFM Probes)
- Fused silica reference sample (nominal MOE 72.9 GPa) or any other reference sample having the MOE value higher than 70 GPa
- TGT1 commercial tip characterization grating (NT-MDT Co.)
- MultiMode 8 atomic force microscope with a Nanoscope V controller and a PeakForce Quantitative Nanomechanical Mapping mode (Bruker Corporation)
Software
- NanoScope analysis (version 1.40r2) (Bruker Corporation)
- MATLAB (version 7) (MathWorks)
Procedure
- Separate leaf midrib samples (0.3-0.4 cm x 0.3-0.4 cm) from the donor plants (Figures 2A-B).
- Immerse the samples into a fixative solution of 5% (v/v) glutaraldehyde (Grade I) in a 0.1 M cacodylate buffer at pH 7.2, and then place them in a vacuum for 15 min at room temperature to remove air from the leaf tissue. Fix the samples in a fixative solution for six hours at 4 °C.
- Dehydrate the samples in an ascending series of ethanol solutions for 20 min each step at room temperature, follow with mixtures of ethanol and xylene at the ratios 3:1, 1:1 and 1:3 (v/v) for 30 min each, ending with pure xylene for 30 min.
- Continue with mixtures of xylene and paraplast plus at the ratios 3:1, 1:1 and 1:3 (v/v) for 30 min each at 42 °C.
- Immerse the samples into pure paraplast plus for 2-3 days and keep them in a laboratory drying chamber at 60 °C. Transfer the samples into freshly prepared paraplast plus twice a day.
- Embed the leaf midrib samples in paraplast plus blocks (Figure 2C).
- Cut leaf midrib cross-sections, 10-15 micrometer-thick, with a microtome.
- Transfer the cross-sections to water droplets on circle glass slides coated with (3-aminopropyl)triethoxy-silane. After relaxation of the sections, carefully remove the water, place the slides on a slide warmer, and allow them to air dry for 2 h at 45 °C.
- Deparaffinize the cross-sections in two successive immersions in xylene for 3 min each, followed by rinsing 3 min each with mixtures of xylene and ethanol at the ratios 3:1, 1:1 and 1:3 (v/v), and complete the process with two successive rinses in 100% ethanol for 3 min each.
- Allow the cross-sections to air dry for 20 min and keep them in sterile Petri dishes.
- Determine the dimensions of tip cantilever from the calibrated light microscopy image, and insert the tip cantilever into the probe holder.
- Calibrate the deflection sensitivity of the tip cantilever on a clean fused silica surface using a PeakForce QNM in air mode. Use a single ramp test. Accept the value to be automatically entered into the deflection sensitivity parameter. (Fused silica is a suitable reference sample for determination of deflection sensitivity of stiff cantilevers, approximately 5 N/m, which are required for the measurement of samples with MOE less than 10 GPa.)
- Use the Nanoscope Thermal Tune function with a simple harmonic oscillator model to obtain the quality factor (Q value) and the resonance frequency (v0 value) of the tip cantilever. Following the Sader’s method of spring constant calibration, enter the dimensions of tip cantilever together with the above values for the quality factor and the resonance frequency, and directly calculate the stiffness of the tip cantilever on this website: http://www.ampc.ms.unimelb.edu.au/afm/calibration.html. Add the calculated value into the spring constant parameter window.
- Image one peak of TGT1 tip grating using a PeakForce QNM, and calculate the tip radius for expected depth. Use the fact that the tip might be approximated by ellipsoid surface. Calculate the fitted ellipsoid below the expected depth using the least square method. If the radii are similar, use the average. In other cases, use another tip or anticipate a non-ideal tip.
- In case of difficulties with the calibration of deflection sensitivity, spring constant or tip radius for a novice user, consult the calibraton chapter of the PeakForce QNM User Guide (available on the websites: http://nanopicolab.cnsi.ucla.edu/pages/publicview/manuals/PEAKFORCE_QNM_USERS_GUIDE-A.pdf or http://www.torontomicrofluidics.ca/cms/manuals/peak_force.pdf).
- Place the circle glass slide containing the sample cross-section onto a steel sample mounting disk using an adhesive pad.
- Place the steel sample mounting disk with the glass slide onto the scanner tube of a microscope.
- Image the tracheary element cell wall fragment within the primary xylem tissue (Figure 2D) by adjusting the peak force setpoint to achieve a sufficient sample deformation of 2 nm.
- Perform the measurement of cell wall topography (i.e. heights), MOE, adhesion, deformation, and energy dissipation at low approach tip velocities (approximately 0.3-0.5 μm/s).
- Import the raw data of MOE into the MATLAB software. The data of MOE must be filtered in order to account for the slippery effect of the tip on the steep cell wall surface. Calculate the height gradients (by means of a function “gradient”) using the height data of surface topography, and also calculate the surface slope (an angle between the gradient vector and the horizontal x, y plane) for each image pixel. Disregard MOE values where the surface slope exceeds the value of 30 degrees, and remove these values from the quantification of MOE. The values for other nanomechanical properties such as adhesion, deformation, and energy dissipation do not seem to be effected by high surface slope. However, to be consistent with the spatial distribution and the statistics of the measured characteristics, we suggest that the filtered data of adhesion, deformation, and energy dissipation be taken into account as well.
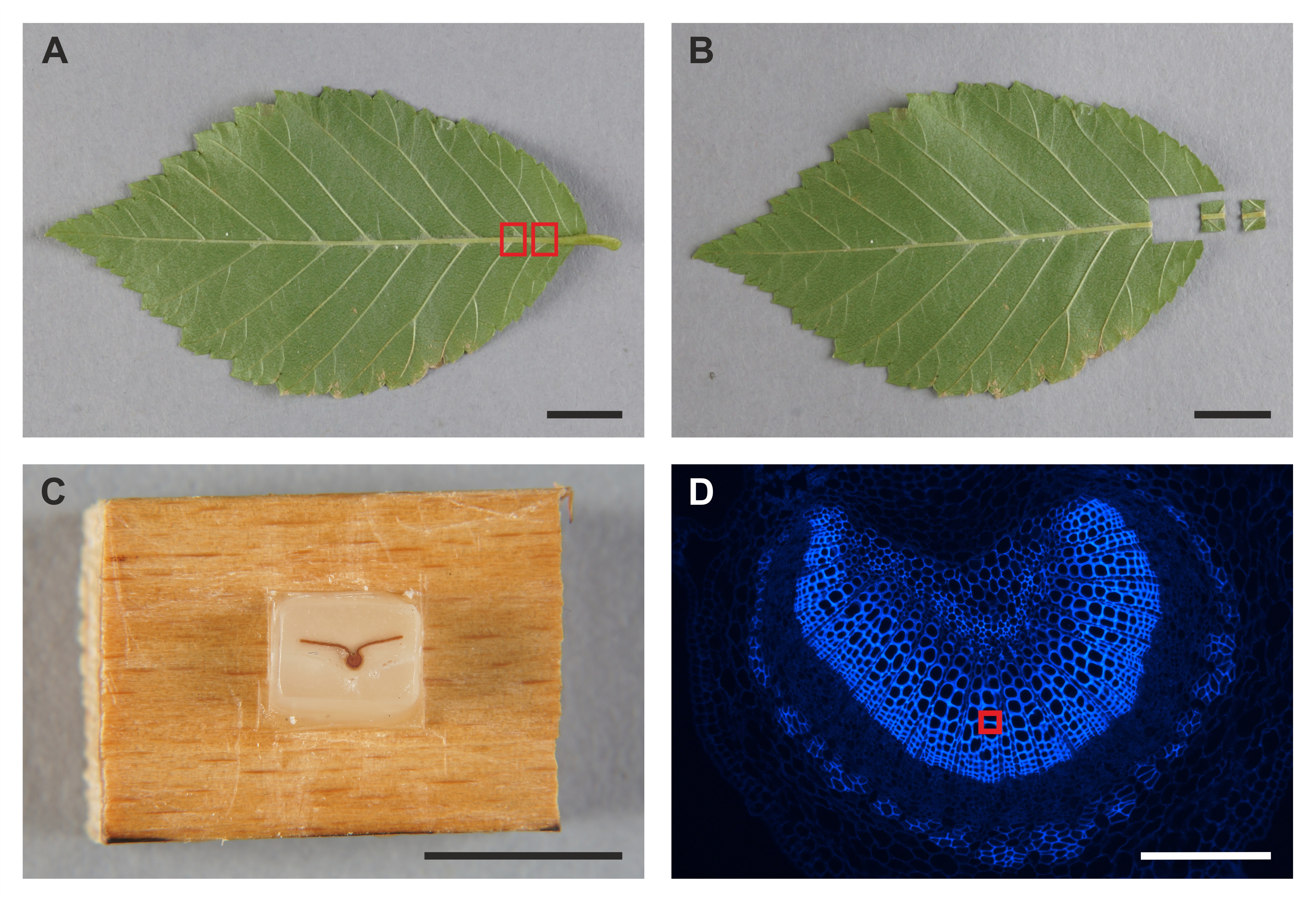
Figure 2. Leaf midrib separation and preparation of a primary xylem tissue for the PeakForce QNM measurement. A. Fully expanded leaf of Ulmus, free of leaf midrib damage. The red boxes indicate the sites of leaf midrib separation. Scale bar = 1 cm. B. Leaf midrib samples separated for the tissue fixation and embedding. Scale bar = 1 cm. C. Leaf midrib sample embedded in a paraplast plus block, ready for microtome sectioning. Scale bar = 1 cm. D. Primary xylem tissue and lignin autofluorescence within tracheary elemet cell walls. The red box indicates the cell wall fragment subjected to the PeakForce QNM measurement. Scale bar = 100 μm
Representative data
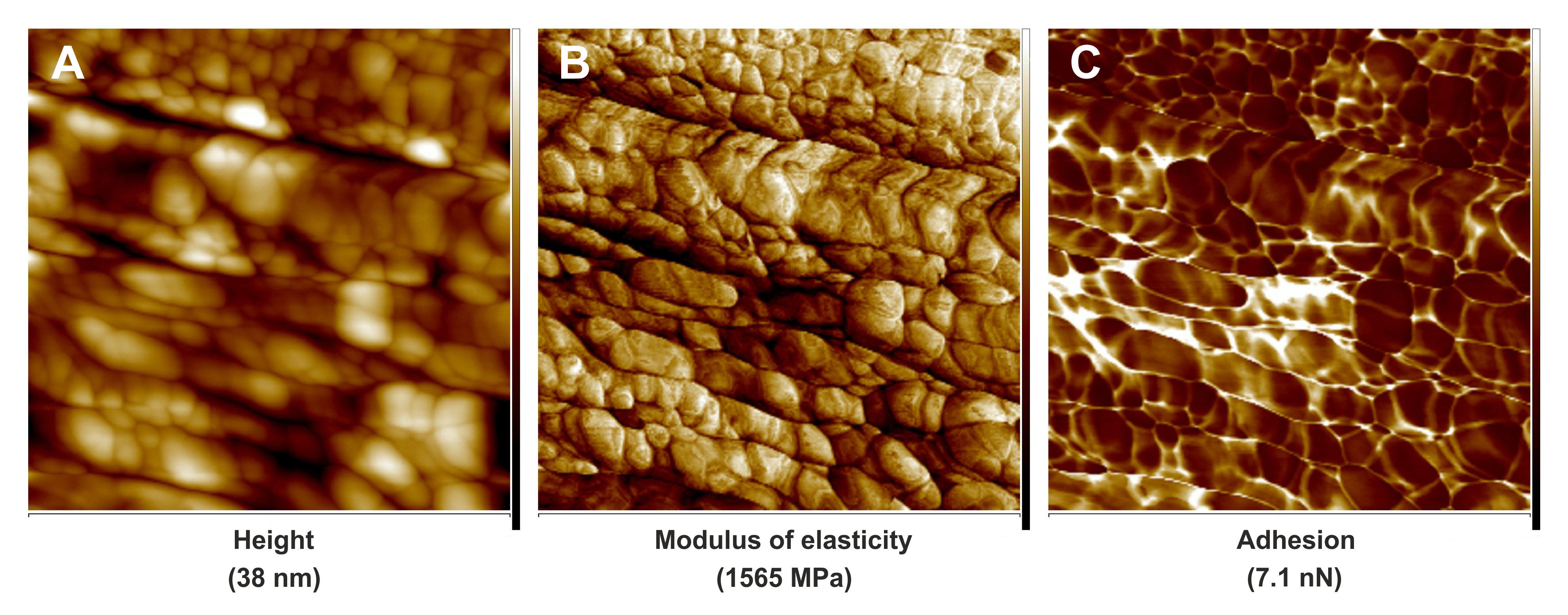
Figure 3. PeakForce QNM images of cell wall fragment surfaces of leaf midrib tracheary elements in the Dutch elm disease-tolerant hybrid 'Dodoens' infected by the pathogenic fungus Ophiostoma novo-ulmi ssp. americana × novo-ulmi. A. Quantitative image for flatten height. B. Quantitative image for modulus of elasticity. C. Quantitative image for adhesion. Scale bars = 1.6 μm
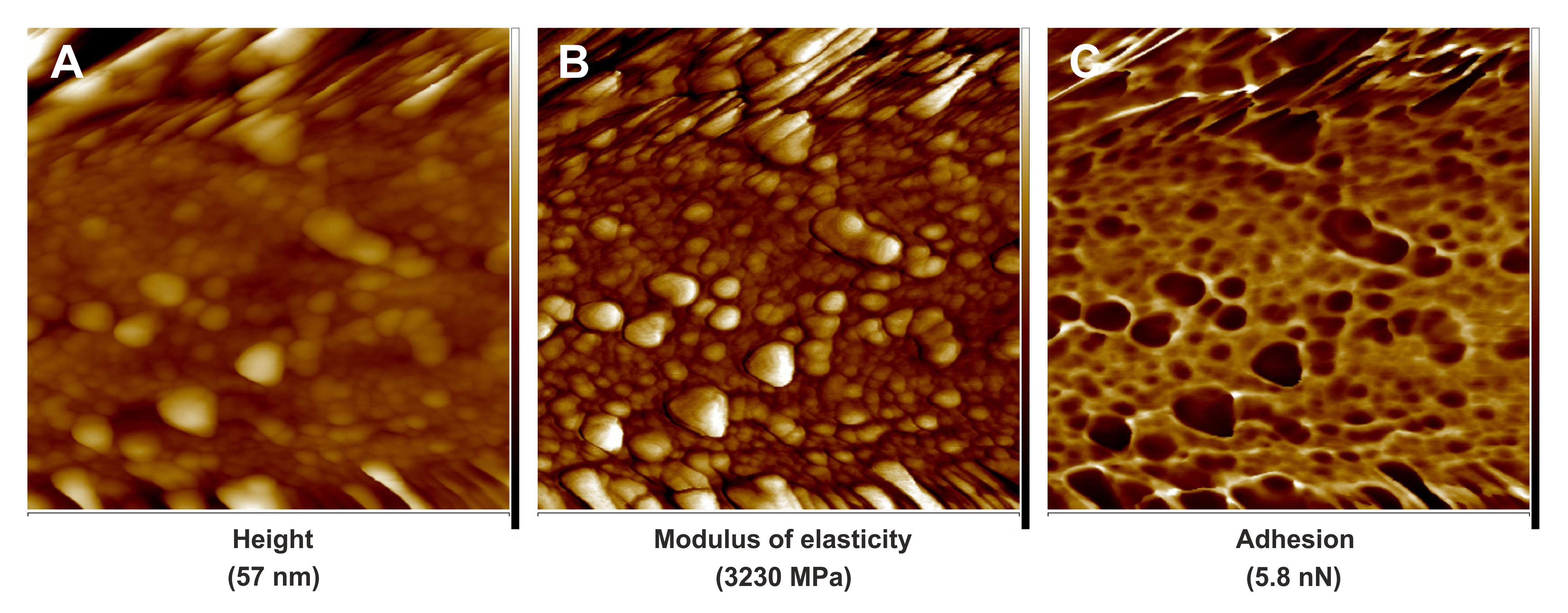
Figure 4. PeakForce QNM images of cell wall fragment surfaces of leaf midrib tracheary elements in the natural hybrid species, Sorbus zuzanae. A. Quantitative image for flatten height. B. Quantitative image for modulus of elasticity. C. Quantitative image for adhesion. Scale bars = 1.1 μm
Recipes
- 0.1 M cacodylate buffer (pH 7.2, 500 ml)
Dissolve 10.7 g sodium cacodylate trihydrate in 400 ml of distilled water
Adjust the pH to 7.2 with HCl
Add distilled water to make a total volume of 500 ml
Handle in the fume cupboard
Stored at room temperature
- Circle glass slides coated with (3-aminopropyl)triethoxy-silane
Dip circle glass slides into acetone for 10 min
Dry in a laboratory drying chamber
Dip slides into 3% (v/v) (3-aminopropyl)triethoxy-silane in acetone for 15 min
Rinse thoroughly in acetone
Dry slides in a laboratory drying chamber
Activate (3-aminopropyl)triethoxy-silane by placing slides into 2.5% (v/v) glutaraldehyde (Grade II) in 1x phosphate buffered saline for 1 h
Wash slides in ultrapure water and air dry
- 1x phosphate buffered saline (1 L)
Dissolve 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4 in 800 ml of distilled water
Adjust pH to 7.4 with HCl
Add distilled water to make a total volume of 1 L
Stored at room temperature
- Glutaraldehyde fixative solution [5% (v/v)]
Prepared by diluting of 25% glutaraldehyde, Grade I in a 0.1 M cacodylate buffer
Work with glutaraldehyde only in a properly operating and certified fume cupboard
Acknowledgments
The authors thank Dr. Olivier Arnould for sharing his MATLAB script for the calculation of tip radius from the TGT1 tip grating images, and Dr. Ingrid Čaňová for her technical assistance. This work was funded by the Slovak scientific grant agency VEGA (1/0132/12). The procedure described above was originally described in Ďurkovič et al. (2013).
References
- Ďurkovič, J., Čaňová, I., Lagaňa, R., Kučerová, V., Moravčík, M., Priwitzer, T., Urban, J., Dvořák, M. and Krajňáková, J. (2013). Leaf trait dissimilarities between Dutch elm hybrids with a contrasting tolerance to Dutch elm disease. Ann Bot 111(2): 215-227.
- Ďurkovič, J., Kardošová, M., Čaňová, I., Lagaňa, R., Priwitzer, T., Chorvát, D., Jr., Cicák, A. and Pichler, V. (2012). Leaf traits in parental and hybrid species of Sorbus (Rosaceae). Am J Bot 99(9): 1489-1500.
- Heu, C., Berquand, A., Elie-Caille, C. and Nicod, L. (2012). Glyphosate-induced stiffening of HaCaT keratinocytes, a Peak Force Tapping study on living cells. J Struct Biol 178(1): 1-7.
- Sweers, K., van der Werf, K., Bennink, M. and Subramaniam, V. (2011). Nanomechanical properties of alpha-synuclein amyloid fibrils: a comparative study by nanoindentation, harmonic force microscopy, and Peakforce QNM. Nanoscale Res Lett 6(1): 270.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ďurkovič, J., Kardošová, M. and Lagaňa, R. (2014). Imaging and Measurement of Nanomechanical Properties within Primary Xylem Cell Walls of Broadleaves. Bio-protocol 4(24): e1360. DOI: 10.21769/BioProtoc.1360.
Category
Plant Science > Plant cell biology > Cell imaging
Cell Biology > Cell structure > Cell adhesion
Cell Biology > Cell imaging > Fixed-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










