- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Stable Transformation of Cyanobacterium Synechocystis sp.
Published: Vol 4, Iss 21, Nov 5, 2014 DOI: 10.21769/BioProtoc.1286 Views: 14157
Reviewed by: Tie LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Fast and Easy Method to Study Ralstonia solanacearum Virulence upon Transient Gene Expression or Gene Silencing in Nicotiana benthamiana Leaves
Wenjia Yu and Alberto P. Macho
Aug 5, 2021 4700 Views

A New Tool for the Flexible Genetic Manipulation of Geobacillus kaustophilus
Ryotaro Amatsu [...] Ken-ichi Yoshida
Sep 5, 2022 1905 Views

Efficient Genetic Transformation and Suicide Plasmid-mediated Genome Editing System for Non-model Microorganism Erwinia persicina
Tingfeng Cheng [...] Lei Zhao
Mar 20, 2024 2442 Views
Abstract
Cyanobacteria are prokaryotes, which perform oxygenic photosynthesis. Among them, the unicellular cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis) is a well characterized model system for studies on oxygenic photosynthesis, light signal transduction etc. Moreover, Synechocystis is applied in biotechnological applications (Desai and Atsumi, 2013). Stable transformation of Synechocytis is achieved via the uptake of DNA and incorporation into the host genome by homologous double recombination. This allows for the generation of gene knock-outs (KO) by replacing the coding sequence of the gene of interest by a KO-cassette (comprising of a selection marker flaked by sequences of the gene of interest) or stable overexpression of certain genes of interest after insertion of a corresponding overexpression cassette at a neutral insertion site on the host genome. Stable transformation of Synechocystis was reported by Grigorieva and Shestakov (1982). Since then, variants of the initial protocol have been applied successfully to transform Synechocystis sp. Here we describe a lab-protocol that was applied successfully for stable transformation of Synechocystis (Schwarzkopf et al., 2014).
Keywords: TransformationMaterials and Reagents
- Synechocystis sp. PCC 6803 wild-type (WT) strain [see Schwarzkopf et al. (2014) for details]
- Antibiotics
- Chloramphenicol (Merck KGaA, catalog number: 2366 )
- Kanamycin sulfat (Carl Roth, catalog number: T832.1 )
- Spectinomycin (Duchefa Biochemie, catalog number: S 0188.0025 )
- Chloramphenicol (Merck KGaA, catalog number: 2366 )
- Phyto agar (Duchefa Biochemie, catalog number: P1003.1000 )
- NaNO3 (Carl Roth, catalog number: 8601.2 )
- K2HPO4 (Carl Roth, catalog number: P749.2 )
- MgSO4.7 H2O (Carl Roth, catalog number: P027.2 )
- CaCl2.2 H2O (Sigma-Aldrich, catalog number: 223506 )
- Citric acid (Carl Roth, catalog number: 1818.1 )
- Ferric ammonium citrate (III+) (Carl Roth, catalog number: CN77.1 )
- EDTA Na2 (Carl Roth, catalog number: 8043.2 )
- Na2CO3 (Sigma-Aldrich, catalog number: S-1641 )
- H3BO3 (Carl Roth, catalog number: 6943.3 )
- MnCl2.4 H2O (Sigma-Aldrich, catalog number: M3634 )
- ZnSO4.7 H2O (Merck KGaA, catalog number: 0 143532 )
- Na MoO4.5 H2O (Carl Roth, catalog number: 0274.3 )
- CuSO4.5 H2O (Carl Roth, catalog number: P025.1 )
- Co(NO3)2.6 H2O (Merck KGaA, catalog number: A834336548 )
- Na2S2O3.5 H2O (Merck KGaA, catalog number: K5023616 )
- BG11 medium (see Recipes)
- BG11 agar plates (see Recipes)
- Common antibiotics used (see Recipes)
Equipment
- Petri dishes (Greiner Bio-one, catalog number: 632180 )
- 2 ml-reaction tubes (Eppendorf)
- Tape (Gotha-VLIES, 10 m x 1.25 cm) (Gothaplast, catalog number: PZN-7105417 )
- Centrifuge (Eppendorf, model: 5810R )
- Shaking incubator with illumination (Sartorius, model: Certomat® BS-T )
- Light shelves provided with light bulbs (NARVA LT 36W/760-010 daylight) (Brand-Erbisdorf)
- Photometer (Pharmacia LKB-Ultrospec III)
- Flow cabinet (Heraeus, HERAsafe, model: HS12 )
Procedure
- All the handling of Synechocystis cells was done in a flow cabinet. Sterile working conditions are critical throughout the whole procedure.
- Synechocystis cells were maintained on BG11 agar plates under continuous illumination (20 μmol/m2/s) at 28 °C and were restreaked in 2-week intervals.
- To generate batch cultures, 25 ml liquid BG11 medium (in a 100-ml-glass beaker) was inoculated with one inoculation loop of Synechocystis cells and cells were grown to an optical density (OD750) of 0.5 to 0.8 (mid-exponential growth) under continuous illumination at a light intensity of 20 μmol/m2/s at 28 °C and permanent agitation (150 rpm). Depending on the inoculation density it takes several days to about one week to reach the indicated OD750 of 0.5 to 0.8.
- 10 ml of cell cultures were centrifuged (5 min, 28 °C, 1,107 x g) and the cell pellet was resuspended in 5 ml BG11 medium and equally distributed to five sterile 2 ml-reaction tubes.
- DNA (3 to 5 μg of circular plasmid DNA) was added and cells were gently mixed by tapping and incubated in darkness at 28 °C over night. To measure the viability and spontaneous mutation frequencies one aliquot was left without plasmid DNA and incubated in darkness at 28 °C over night.
- Following dark incubation, 200 μl of the cell cultures were plated on BG11 agar plates (94 x 16 mm, see “Materials” for details) lacking antibiotics and incubated in continuous light at a light intensity of 20 μmol/m2/s at 28 °C.
- After 2 days of incubation under constant light, the agar plates were supplied with antibiotics at a concentration of 15 μg/ml. Therefore, the agar was lifted with a sterile spatula and 1 ml of the appropriate antibiotic solution was applied to the bottom of the petri dish (Figure 1). In case several different antibiotics have to be applied prepare an antibiotic mixture containing each antibiotic combined in 1 ml of total volume.
- Plates were sealed with tape to avoid drying-out and further incubated under continuous illumination at a light intensity of 20 μmol/m2/s at 28 °C. Colonies appeared after 2 to 3 weeks.
- To achieve complete segregation, single colonies were picked and streaked on BG11 plates containing the appropriate antibiotics (see step 10 for appropriate antibiotic concentrations). When cells were gown (about one to two weeks) cells were streaked again on new BG11 plates containing the appropriate antibiotics. This successive streak purification was repeated at least four rounds (in total) to ensure segregation.
- To check for positive genomic integration of the construct PCR on genomic DNA was performed. It might happen that complete segregation cannot be achieved even after several rounds of successive streak purification. This might happen when a gene knock-out is attempted of a gene, which is indispensable for viability of the cells. In such a case anti-sense approaches or overexpression of dominant negative forms might be applied to interfere with the function of the gene of interest.
- Positive clones were maintained on BG11 agar plates containing appropriate antibiotics under continuous illumination (20 μmol/m2/s) and were restreaked in 2-week intervals. Common antibiotics used are chloramphenicol at 8 μg/ml, spectinomycin at 10 μg/ml, kanamycin at 10 μg/ml.
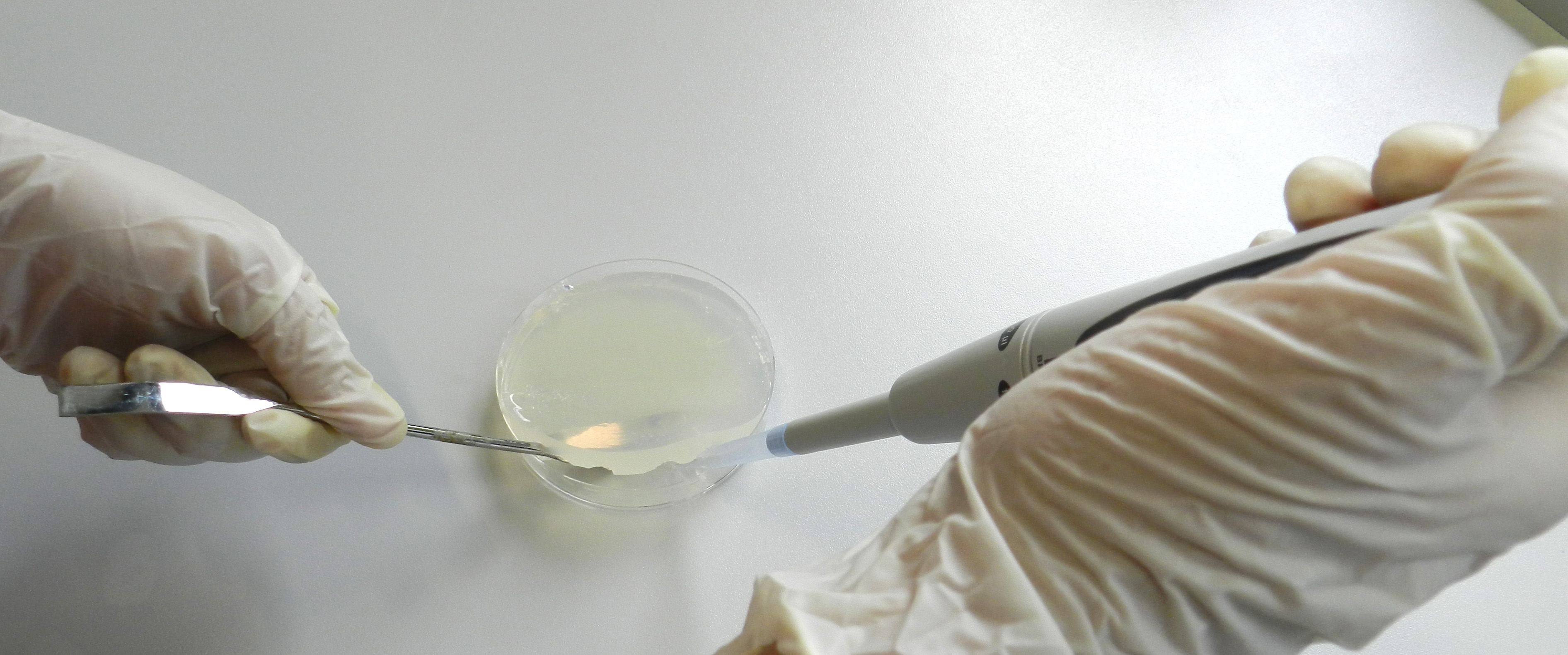
Figure 1. Applying antibiotics beneath the agar layer. A sterile curved spatula was used to lift the agar layer and antibiotic solutions were applied to the bottom of the petri dish.
Notes
- Transformation efficiency may vary, depending on the cell status and the construct used for transformation and commonly result in 10 to 50 clones per approach.
Recipes
- BG11 medium (Stanier et al., 1971)
BG11 medium was prepared in 10x concentration, autoclaved at 121 °C for 12 min and stored at 4 °C. Before use, 890 ml H2Odest, 100 ml 10x BG11 and 10 ml of the trace metal solution were combined to yield 1L of 1x BG11. BG11 medium was aliquoted in culture vessels, autoclaved and stored at 4 °C.
10x BG11 medium:
NaNO3 15 g
K2HPO4 0.4 g
MgSO4.7 H2O 0.75 g
CaCl2.2 H2O 0.36 g
Citric acid 0.06 g
Ferric ammonium citrate (III+) 0.06 g
EDTA Na2 0.01 g
Na2CO3 0.2 g
Add H2Odest 1 L
Autoclaved and stored at 4 °C
Trace metal solution per litre:
H3BO3 2.86 g
MnCl2.4 H2O 1.81 g
ZnSO4.7 H2O 0.22 g
Na MoO4.5 H2O 0.39 g
CuSO4.5 H2O 0.08 g
Co(NO3)2.6 H2O 0.05 g
- BG11 agar plates
For solid medium 1x BG11 was supplemented with 1.5 % (w/v) phyto agar, 0.18 % (w/v) sodium thiosulfate (Na2S2O3.5 H2O; added for better solidification of the agar), autoclaved and poured in petri-dishes (approximately 30 ml per dish). Where appropriate, filter-sterilized antibiotics were added to the medium.
- Common antibiotics used
Chloramphenicol at 8 μg/ml
Spectinomycin at 10 μg/ml
Kanamycin at 10 μg/ml
Acknowledgments
Stable transformation of Synechocystis was reported by Grigorieva and Shestakov (1982). Experimental work was supported by Technische Universität München (TUM) core funding.
References
- Desai, S. H. and Atsumi, S. (2013). Photosynthetic approaches to chemical biotechnology. Curr Opin Biotechnol 24(6): 1031-1036.
- Grigorieva, G. and Shestakov, S. (1982). Transformation in the cyanobacterium Synechocystis sp. 6803. FEMS Microbiol Lett 13(4): 367-370.
- Schwarzkopf, M., Yoo, Y. C., Huckelhoven, R., Park, Y. M. and Proels, R. K. (2014). Cyanobacterial phytochrome2 regulates the heterotrophic metabolism and has a function in the heat and high-light stress response. Plant Physiol 164(4): 2157-2166.
- Stanier, R. Y., Kunisawa, R., Mandel, M. and Cohen-Bazire, G. (1971). Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol Rev 35(2): 171-205.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Proels, R. K. (2014). Stable Transformation of Cyanobacterium Synechocystis sp.. Bio-protocol 4(21): e1286. DOI: 10.21769/BioProtoc.1286.
- Schwarzkopf, M., Yoo, Y. C., Huckelhoven, R., Park, Y. M. and Proels, R. K. (2014). Cyanobacterial phytochrome2 regulates the heterotrophic metabolism and has a function in the heat and high-light stress response. Plant Physiol 164(4): 2157-2166.
Category
Microbiology > Microbial genetics > Transformation
Molecular Biology > DNA > Transformation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









