- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Protocol to Measure the Extent of Cell-to-cell Movement of RNA Viruses in Planta
Published: Vol 4, Iss 20, Oct 20, 2014 DOI: 10.21769/BioProtoc.1269 Views: 8368
Reviewed by: Hiromasa SaitohTie LiuArsalan Daudi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of the Composition Dynamics of a Maize Root-associated Simplified Bacterial Community and Evaluation of Its Biological Control Effect
Ben Niu and Roberto Kolter
Jun 20, 2018 10502 Views

Tracking Root Interactions System (TRIS) Experiment and Quality Control
Hassan Massalha [...] Asaph Aharoni
Apr 20, 2019 7345 Views

A Quick Method for Screening Biocontrol Efficacy of Bacterial Isolates against Bacterial Wilt Pathogen Ralstonia solanacearum in Tomato
Heena Agarwal [...] Niraj Agarwala
Nov 20, 2020 5559 Views
Abstract
Here, we present a simple and rapid protocol to measure the extent of cell-to-cell movement of RNA viruses in planta. To do that, the green fluorescent protein (GFP) gene was incorporated into the genome of Melon necrotic spot virus (MNSV) as a coat protein (CP) fusion protein using the Thosea asigna virus 2A catalytic peptide (TaV 2a) (Serra-Soriano et al., 2014). TaV 2a allows the co-translational cleavage of the fusion protein resulting in the independent expression of both proteins (Kim et al., 2011). Viral infection was initiated by agro-infiltration of Cucumis melo leaves. At 6-7 days post-infiltration, fluorescent infection foci images were taken with a fluorescent stereo microscope and infection areas were measured using FIJI software.
Keywords: Plant virusMaterials and Reagents
- 3-4 weeks old Cucumis melo L. subsp. melo cv. Galia plants
- Agrobacterium tumefaciens (A. tumefaciens) strain C58C1, or similar, transformed with the binary vector harboring the GFP-tagged viral genome
In our case, pMNSV(Al)/GFP encoding GFP-tagged Melon necrotic spot virus (MNSV) genome under the control of the Cauliflower mosaic virus (CaMV) 35S promoter and the potato proteinase inhibitor terminator (PoPit) (Figure 1).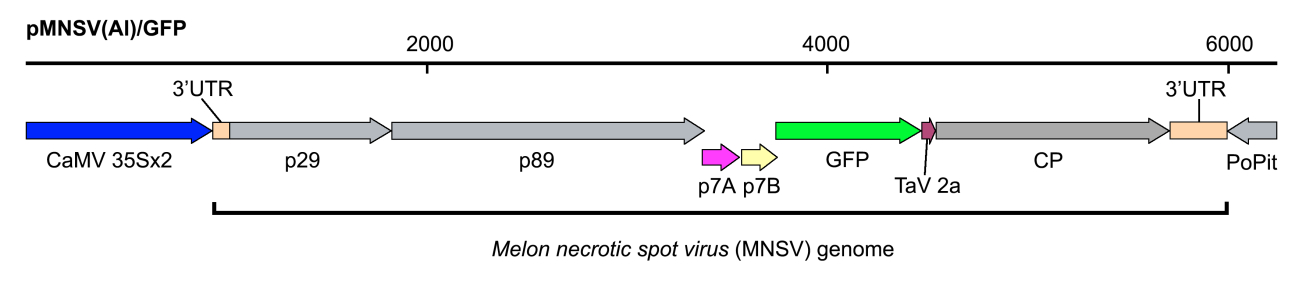
Figure 1. Schematic representation of the recombinant infectious clone pMNSV(Al)/GFP used in this assay - Yeast extract (Difco, catalog number: 212750 )
- Tryptone (Difco, catalog number: 211705 )
- Sodium chloride (NaCl) (Panreac Applichem, catalog number: 131659 )
- Acetosyringone (Sigma-Aldrich, catalog number: D134406 )
- MES (Sigma-Aldrich, catalog number: M8250 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M9272 )
- Antibiotics
- LB medium (see Recipes)
- Agrobacterium infiltration buffer (see Recipes)
Equipment
- 28 °C growing chamber
- 15 ml culture tubes
- Swinging centrifuge rotor for 15 ml tubes
- 1.5 ml tubes (standard Eppendorf tubes or similar)
- BioPhotometer plus (Eppendorf, catalog number: 6132000008 )
- 1 ml syringes without needle
- Plant growing chamber
- Leica MZ12 fluorescent stereo microscope
Software
- Adobe Photoshop CS5 or higher
- ImageJ, FIJI or similar software
- MS Excel
Procedure
- Viral inoculation by agro-infiltration
- Inoculate a single colony of A. tumefaciens transformed with pMNSV(Al)/GFP in 5 ml of LB medium with Kanamycin (50 µg/ml) and Rifampicin (50 µg/ml). Incubate the culture overnight (about 12-16 h) at 28-30 °C with vigorous shaking.
- Measure the A600 of the culture.
- Collect the bacteria by slow-speed centrifugation (2,000 x g, 15 min) in a swinging rotor. Remove the supernatant.
- Resuspend the pellet with Agrobacterium infiltration buffer. Since some viruses replicate and then move faster than others, the final A600 need to be adjusted depending on the virus. Large inoculum of a fast-moving virus may generate a high number of viral infection foci so close between them that they could be quickly merged. Therefore the resulting average size of the infection foci could be overestimated. If this occurs, reduce the final A600 until you get infection foci physically separated from each other. If, instead, only few infection foci are observed, the final A600 must be increased.
- Leave at room temperature for 2 h before infiltration.
- Infiltrate the cultures with 1 ml syringe into the lower side of the leaves. Use 3-4 week-old Cucumis melo plants and avoid non fully-expanded leaves. Use your fingertip to apply gentle counter pressure to the other side of the leaf. A fully infiltrated part of the leaf gives a water-soaked appearance.
- Label de infiltrated leaves with a ring of adhesive one-sided tape placed around the petiole.
- Keep the plants in growth chambers in 16 h light at 25 °C and 8 h dark at 22 °C.
- Inoculate a single colony of A. tumefaciens transformed with pMNSV(Al)/GFP in 5 ml of LB medium with Kanamycin (50 µg/ml) and Rifampicin (50 µg/ml). Incubate the culture overnight (about 12-16 h) at 28-30 °C with vigorous shaking.
- Image acquisition
- Several days after infiltration check the appearance of fluorescent infection foci with a Leica MZ12 fluorescent stereo microscope or similar device. The exact time will depend on which virus is assayed (5-8 days after infiltration).
- Place the melon leaves beneath the objective (Planachromatic 0.5x) and engage the lowest magnification factor (0.71x)
- Select GFP1 filter set [excitation filter 425/60 nm (395-455 nm), barrier filter 480 nm].
- Look into the eyepieces and bring the fluorescent foci into focus using the rotatory knob.
- Correct the values for "exposure", "gain" and "gamma" until you obtain the desired results for the image and capture it. Don’t forget to add a scale bar.
- Several days after infiltration check the appearance of fluorescent infection foci with a Leica MZ12 fluorescent stereo microscope or similar device. The exact time will depend on which virus is assayed (5-8 days after infiltration).
- Image analysis
- Open the image with Adobe Photoshop CS5 or higher.
- If necessary, enhance the image brightness and contrast until infection foci have well-defined borders.
- Use the magic wand selection tool to select the infection foci. This tool selects similarly colored non-contiguous areas. If the magic wand selects less area than you want, increase the tolerance setting.
- Fill selection with black color by selecting “Fill” in the “Edit” menu.
- Invert selection by choosing “Invert” in the “Select” menu and fill with white color. If you have infection foci that have merged together separate them by drawing a white line were it feels the division should be. Save the resulting black and white image (Figure 2).
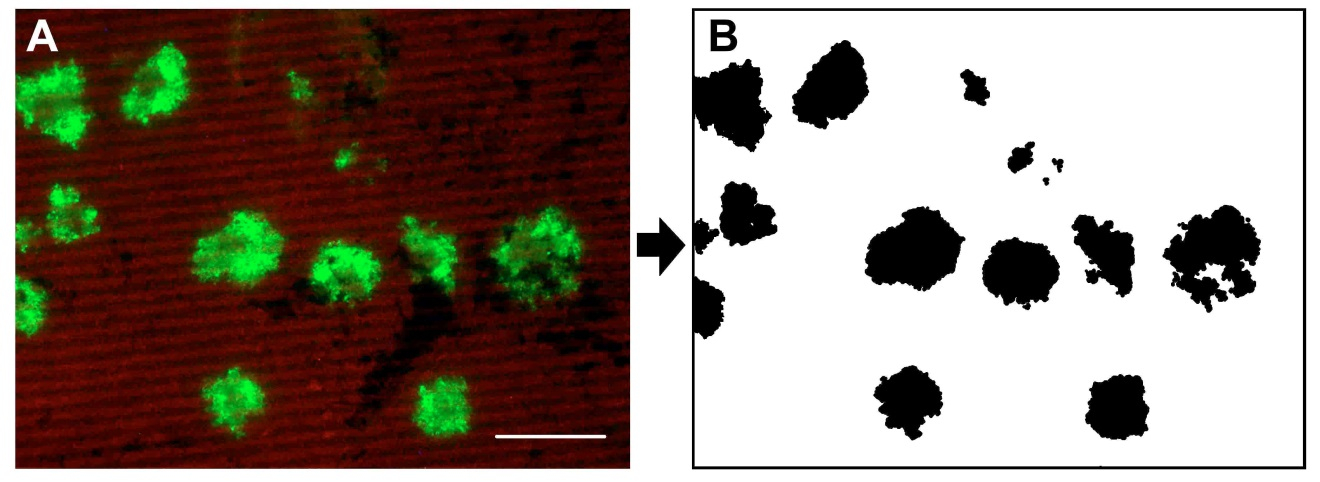
Figure 2. Images of after B and before A the Adobe Photoshop editing - Open the black and white image with ImageJ.
- Select “Binary” in the “Process” menu and “Make binary”.
- Check the spatial calibration of the image. If the image is calibrated, the number of pixels in each dimension and dimensions with a unit of length will be visible at the top of the image window. If the image is not calibrated, only the number of pixels will be written and you will need to set the dimension of the image manually. Then, open an image with a scale bar and using the "Straight line" tool, draw a line along the scale bar. In the “Analyze” menu select “Set scale”. A dialog box will open and the distance in pixels will be automatically adjusted. By choosing global, you set this calibration for all images you open during the ImageJ session.
- Select “Analyze particles” in the “Analyze” menu. In the new dialog box, set size range by trial and error to exclude tiny background particles. Choose “Exclude on edges” option to avoid counting partial infection foci at the edges of the image. Check “Include holes” if you want it to fill in holes and include that area in the measurement. Under “Show” select “Overlay outlines” to number the infection foci that are measured. These numbers correspond to data for individual particles that will be listed in the “Results” window if you check the “Display results” and click “Ok”.
- Save results as MS Excel file and calculate the average size of infection foci in MS Excel.
- Open the image with Adobe Photoshop CS5 or higher.
Recipes
- LB medium
Mix 5 g of yeast extract with 10 g of tryptone, 10 g of NaCl
Add dH2O to 1,000 ml - Agrobacterium infiltration buffer
10 mM MES (pH 5.6)
10 mM MgCl2
150 µM acetosyringone
Acknowledgments
This work was funded by grant BIO2011-25018 from the Spanish Ministerio de Economia y Competitividad and by Prometeo Program GV2011/003 from the Generalitat Valenciana. J.A.N. and M.S. are the recipients of a postdoctoral contract and a PhD fellowship from the Ministerio de Educacion y Ciencia of Spain.
References
- Kim, J. H., Lee, S. R., Li, L. H., Park, H. J., Park, J. H., Lee, K. Y., Kim, M. K., Shin, B. A. and Choi, S. Y. (2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6(4): e18556.
- Serra-Soriano, M., Pallas, V. and Navarro, J. A. (2014). A model for transport of a viral membrane protein through the early secretory pathway: minimal sequence and endoplasmic reticulum lateral mobility requirements. Plant J 77(6): 863-879.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Navarro, J. A., Serra-Soriano, M. and Pallás, V. (2014). A Protocol to Measure the Extent of Cell-to-cell Movement of RNA Viruses in Planta. Bio-protocol 4(20): e1269. DOI: 10.21769/BioProtoc.1269.
Category
Plant Science > Plant immunity > Perception and signaling
Plant Science > Plant immunity > Disease bioassay
Microbiology > Microbe-host interactions > In vivo model > Plant
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









