- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Centrifuge Microscopy to Analyze the Sedimentary Movements of Amyloplasts
Published: Vol 4, Iss 17, Sep 5, 2014 DOI: 10.21769/BioProtoc.1229 Views: 9423
Reviewed by: Arsalan DaudiTie Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views

A Simple Protocol for Periodic Live Cell Observation of Flagellate Stages in the Lichen Alga Trebouxia
Enrico Boccato [...] Mauro Tretiach
Jan 20, 2026 188 Views
Abstract
A centrifuge microscope (CMS) functionally consists of a centrifuge producing a centrifugal force (hypergravity condition) and a microscope making an enlarged image of an object. This combination of equipment allows live-cell imaging during centrifugation. We have developed a new CMS (NSK Ltd.) to observe movements of the plant organelles such as amyloplasts, under hypergravity conditions (Toyota et al., 2013). This CMS is distinct from previously designed CMSs in terms of spatio-temporal resolution, ease of use and compactness. Here, we show a quick protocol to prepare a specimen of Arabidopsis inflorescence stem, use the CMS, obtain imaging data and analyze them using a single tracking method.
Materials and Reagents
- Arabidopsis thaliana inflorescence stems
- MS salt mixture (Wako Pure Chemical Industries, catalog number: 392-00591 )
- 1% (w/v) sucrose
- 0.05% (w/v) MES
- 0.1% (w/v) agar
- Growth media (see Recipes)
Equipment
- Fine tweezers
- Scissors
- Razor blade (Electron Microscopy Sciences, catalog number: 72000 )
- Kimwipes
- Aluminum chamber (custom built) (NSK Ltd.)
- Silicone rubber (thickness: 0.5 mm) (AS ONE Corporation, catalog number: 6-611-01 )
- Round cover glass (diameter: 12 mm) (Matsunami Glass, catalog number: CO12001 )
- CMS system (Figure 1, not commercially available) (NSK Ltd., http://www.nsk.com/)
CMS is a newly designed compact centrifuge microscope, 30 cm in height and 20 cm in diameter. CMS consists of a direct-drive motor (NSK Ltd., MEGATORQUE MOTOR™, model: M-PS1006KN002 ) and optics including a 50x objective lens with a working distance of 18 mm (SLMPLN 50x, 0.35 NA, OLYMPUS), LED light source (SCHOTT MORITEX Corporation, model: MEBL-CW25 ) and a CCD camera (SENSOR TECHNOLOGY, model: STC172C ).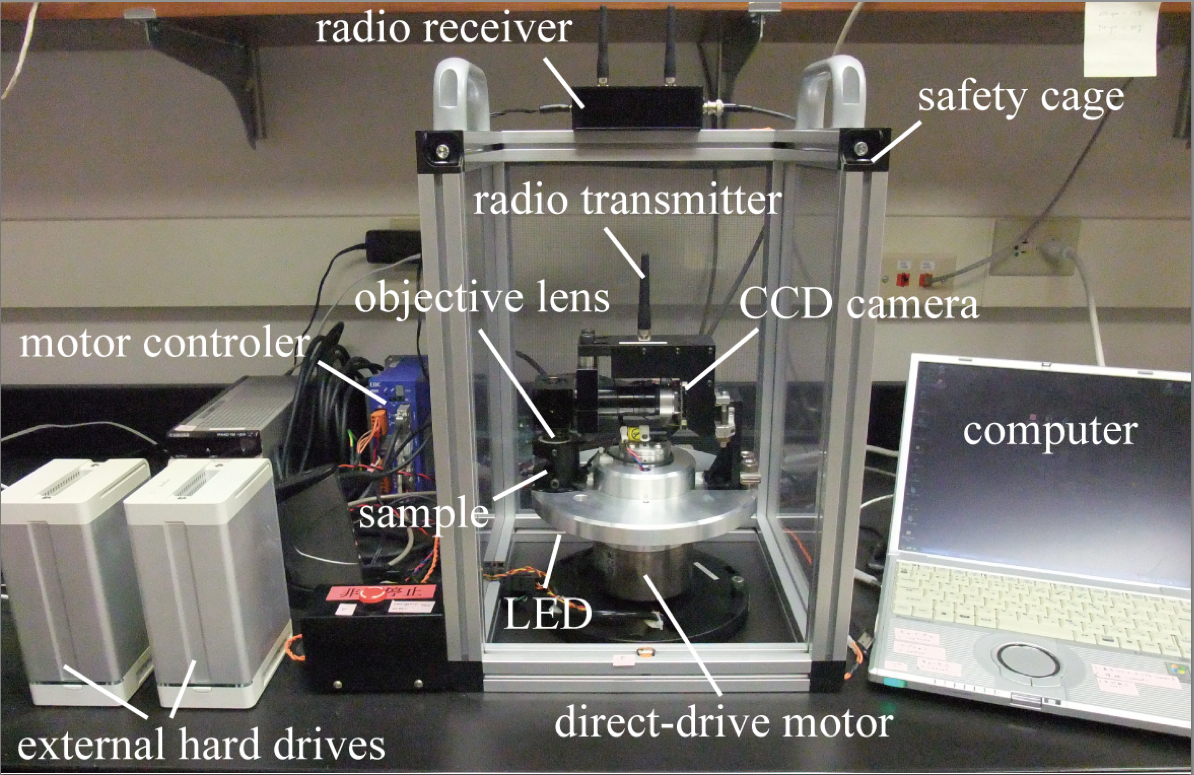
Figure 1. Overview of the CMS system - Windows computer [minimum computer requirements: Windows® XP or later, Pentium M 778 (1.6 GHz), RAM 1024 MB or higher]
Note: To install the software below, Windows computers are highly recommended.
Software
- MEGATORQUE MOTOR™ controller (EDC megaterm software) (NSK Ltd., http://www.nsk.com/)
Note: This is free software to control the motor and is available only for Windows. - Video capture software (COREL, http://www.corel.com/)
Note: You can use any video capture soft/hardware that converts analog video signal into digital signal and stores the data in a computer. - G-Track spot-tracking software (G-Angstrom, http://www.g-angstrom.com/eng/products/index.php)
Note: This is a piece of commercial software to trace fluorescence/bright spots and available only for Windows.
Procedure
- Excise an approximately 1-cm-long segment of an inflorescence stem at 1-2 cm from the apex of the primary stem.
- Place the stem segment onto a hand-made stem holder and slide a razor blade to split the stem longitudinally (Figure 2) (Saito et al., 2005; Nakamura et al., 2011).
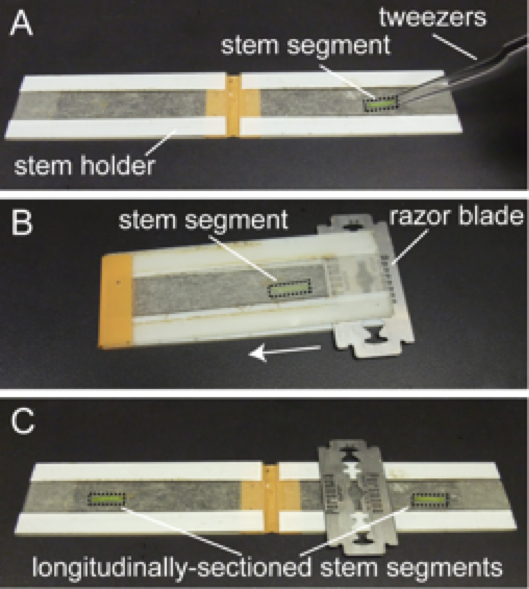
Figure 2. Making longitudinal sections of Arabidopsis inflorescence stems. A. Place the stem segment onto a hand-made stem holder. B. Close the holder and slide a razor blade in the direction of the arrow. C. Open the holder and retrieve the longitudinally-sectioned stem segments. - Drop a small amount of growth medium onto the sectioned side of the stem to prevent the tissue from drying out.
- Keep the sectioned side up and put this segment into a slit in a round silicone rubber sheet (0.5 mm deep) on the bottom glass of an aluminum chamber (Toyota et al., 2013).
- Pour growth medium into this slit, put a cover glass onto the silicone rubber and remove spilt growth media with Kimwipes.
- Mount the aluminum chamber in a holder under an objective lens of the CMS.
- Turn on the LED light to illuminate the specimen and the CCD camera to acquire images.
- Acquired bright-field images are transmitted through a radio system and shown on the video capture software in the computer.
- Adjust focus and field of view in the CMS while monitoring the computer.
- Set a centrifugal acceleration between 0 to 33 x g in the MEGATORQUE MOTOR™ controller of the computer. In case of wild-type Arabidopsis stems, maximum gravitropic responses are seen at 10 x g.
- Start video capture and run MEGATORQUE MOTOR™ (Videos 1 and 2).Video 1. Side view of the CMS during operationVideo 2. Top view of the CMS during operation
- Monitor real-time images (30 frames per sec) on the computer during centrifugation.
- Stop the motor and video capture and save the images as an avi file in the computer.
- Open this file in G-Track spot-tracking software.
- This software automatically recognizes many white or black spots (amyloplasts) and traces them (Video 3; Figure 2). If necessary, you can modify image parameters such as gain or contrast, and invert brightness.Video 3. Single-particle tracking analysis of amyloplast movement during centrifugation at 10 x g (Toyota et al., 2013). Most amyloplasts are automatically recognized by the G-Track spot-tracking software and traced while they are recognized as white or black spots. Please note that this software does not precisely recognize an amyloplast (spot) with weak contrast nor aggregated amyloplasts. Video duration = 152 s (8 x speed).
- Run the tracking program. You can automatically get data [i.e., mean square displacement (MSD) of an amyloplast, Table 1] and then calculate velocity and displacement.
- Export the data to Excel/CSV file (Table 1).
Representative data
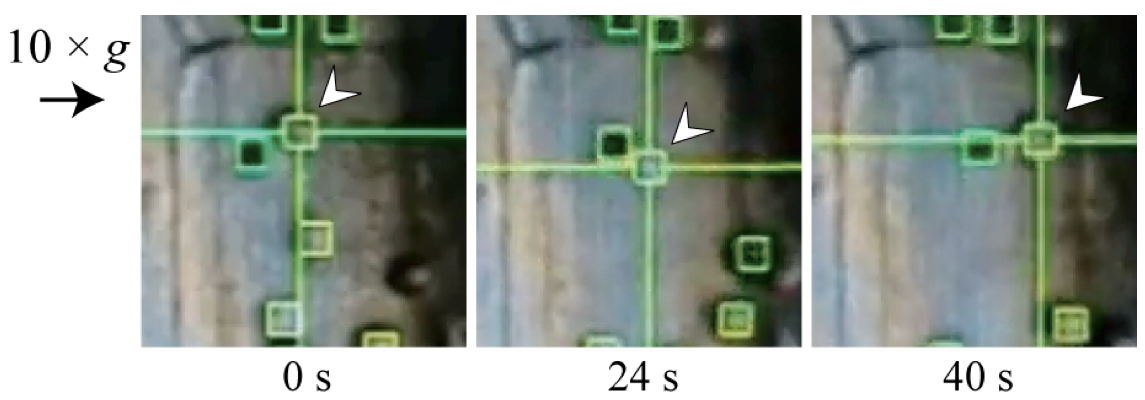
Figure 3. Representative tracking image (Toyota et al., 2013). Movement of an amyloplast (arrow head) is successfully traced by the G-Track spot-tracking software during centrifugation.
Table 1. Mean square desplacement (MSD) of the amylplast for 1 sec of centrifugation. MSD of the amyloplast traced in Figure 3 is automatically calculated by the tracking program. X, Y and 2D denote movement in the horizontal (10 x g) and vertical directions and in a two-dimensional (2D) plane, respectively. For downloading data, please click the image below.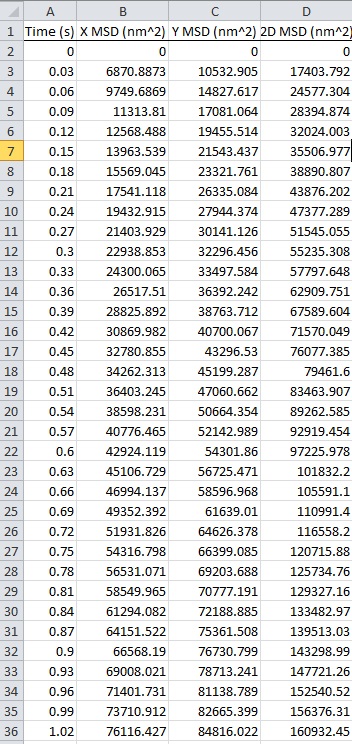
Recipes
- Growth media (pH 5.1)
1x MS salts
1% (w/v) sucrose
0.05% (w/v) MES
0.1% (w/v) agar
Acknowledgments
We thank Professor T. Mimura (Kobe University), Professor Y. Yoshimoto (Kansai Medical University) and Professor T. Shimmen (University of Hyogo) for valuable information about centrifuge microscopes and Y. Ishibashi for technical assistance. This work was supported in part by TOYOBO Biotechnology Foundation (to M. Toyota); Grant-in-Aid for JSPS Fellows (to M. Toyota), for JSPS Fellows for Research Abroad (to M. Toyota) and for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (16085205 to M.T.M.); and grants from the Bioarchitect Project of RIKEN (to M.T.M.) and PREST (to M. Toyota and M.T.M.).
References
- Nakamura, M., Toyota, M., Tasaka, M. and Morita, M. T. (2011). An Arabidopsis E3 ligase, SHOOT GRAVITROPISM9, modulates the interaction between statoliths and F-actin in gravity sensing. Plant Cell 23(5): 1830-1848.
- Saito, C., Morita, M. T., Kato, T. and Tasaka, M. (2005). Amyloplasts and vacuolar membrane dynamics in the living graviperceptive cell of the Arabidopsis inflorescence stem. Plant Cell 17(2): 548-558.
- Toyota, M., Ikeda, N., Sawai‐Toyota, S., Kato, T., Gilroy, S., Tasaka, M. and Morita, M. T. (2013). Amyloplast displacement is necessary for gravisensing in Arabidopsis shoots as revealed by a centrifuge microscope. Plant J 76(4): 648-660.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Toyota, M., Ikeda, N., Tasaka, M. and Morita, M. T. (2014). Centrifuge Microscopy to Analyze the Sedimentary Movements of Amyloplasts. Bio-protocol 4(17): e1229. DOI: 10.21769/BioProtoc.1229.
Category
Plant Science > Plant cell biology > Cell imaging
Plant Science > Plant cell biology > Tissue analysis
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










