- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Tumor-infiltrating Lymphocytes Following CD45 Enrichment
Published: Vol 4, Iss 16, Aug 20, 2014 DOI: 10.21769/BioProtoc.1218 Views: 27131
Reviewed by: Isabel Cristiane da SilvaHongLok LungAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Murine Alveolar Type II Epithelial Cells
Fan Sun [...] Zhaoxia Qu
May 20, 2017 13116 Views

Soft Agar Colony Formation Assay as a Hallmark of Carcinogenesis
Feng Du [...] Daiming Fan
Jun 20, 2017 30592 Views

A Fast and Reliable Method to Generate Pure, Single Cell-derived Clones of Mammalian Cells
Zhe Han [...] Varun Kumar
Aug 20, 2022 4105 Views
Abstract
Measuring antigen-specific T cell responses in the blood and lymphoid organs of vaccinated mice can give us a useful indication of the potency of a vaccine formulation. Unfortunately, systemic or even localized lymphoid T cell responses are not always predictive of the ability of a vaccine to induce tumor protection. Measuring the antigen-specific T cell response within the tumor infiltrating lymphocytes is a more accurate indicator or vaccine efficacy. However, multi-parameter flow cytometric analysis of T cells isolated from tumor tissue can be quite challenging due to the over-whelming number of tumor cells present in relation to the tumor infiltrating lymphocytes (TIL) and to problems associated to the large and adhesive nature of many tumor cell. Here we take advantage of a pre-flow separation of CD45+ leukocytes from the tumor tissue using the MACS magnetic cell sorting system, resulting in a much cleaner cell preparation with which to proceed to flow cytometric staining and analysis.
Keywords: TILMaterials and Reagents
- Cell lines
- The B16.OVA melanoma cell line (Fidler, 1975; Schuler et al., 2008) maintained in cDMEM supplemented with G418-sulphate (geneticin selective antibiotic) (at 1 mg/ml)
Note: Adherent cells detached using 0.25 % Trypsin-EDTA.
- The B16.OVA melanoma cell line (Fidler, 1975; Schuler et al., 2008) maintained in cDMEM supplemented with G418-sulphate (geneticin selective antibiotic) (at 1 mg/ml)
- Mice
- All mice used in this protocol were between 6 and 12 weeks of age and were sex and age matched for each individual experiment. Three mice were used per group in each experiment.
- CD45.1 congenic (B6.SJL-PtprcaPep3b/BoyJArc), bred in-house at the SPF Animal Facility of the UNIL
- OT-I mice (Hogquist et al., 1994)
- OT-IIxFoxp3-eGFP mice [referred to as OT-II (Barnden et al., 1998, Wang et al., 2008)] bred in-house at the SPF Animal Facility of the UNIL
- All mice used in this protocol were between 6 and 12 weeks of age and were sex and age matched for each individual experiment. Three mice were used per group in each experiment.
- Antibodies
- Va2 and Vb5.1/5.2 antibodies (BD Biosciences)
- CD45 PE antibody (BD Biosciences)
- CD45.1 APC-eFluor780 (eBioscience)
- CD45.2 pacific blue (eBioscience)
- CD8 eFluor700 (eBioscience)
- CD4 PE-Texas red (eBioscience)
- 2.4G2 (Anti-FcgRII monoclonal antibody)
- Va2 and Vb5.1/5.2 antibodies (BD Biosciences)
- Buffers and media
- Dulbecco’s modified Eagle’s medium (DMEM), high glucose, GlutaMAX™ supplement (Life Technologies, Gibco®, catalog number: 10566-016 )
- 1 M HEPES (Life Technologies, Gibco®, catalog number: 15630-080 )
- Penicillin-streptomycin (5,000 U/ml) (Life Technologies, Gibco®, catalog number: 15070063 )
- Fœtal bovine serum (FBS). (performance sera with low endotoxin : qualified, US origin) (Life Technologies, catalog number: 26140 or similar)
- Geneticin® selective antibiotic (G418 Sulfate, 50 mg/ml) (Life Technologies, Gibco®, catalog number: 15630-080)
- 0.25% Trypsin-EDTA (1x) (phenol red) (Life Technologies, InvitrogenTM, catalog number: 25200056 )
- Phosphate buffered saline (PBS) (Laboratorium Dr. Bichsel AG)
- Collagenase, Type I (Life Technologies, Gibco®, catalog number: 17018-029 )
- DNAse I (Roche Diagnostics, catalog numner: 0 4536282001 )
- UltraPure™ 0.5 M EDTA (pH 8.0) (Life Technologies, InvitrogenTM, catalog number: 15575-020 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A9418 )
- Complete Dulbecco’s modified Eagle’s medium (cDMEM) (see Recipes)
- Tumor digesting buffer (see Recipes)
- Flow Buffer (see Recipes)
- MACS Buffer (see Recipes)
- Dulbecco’s modified Eagle’s medium (DMEM), high glucose, GlutaMAX™ supplement (Life Technologies, Gibco®, catalog number: 10566-016 )
- Peptides
- OVA257-264 and OVA323-339 peptides
Note: Peptides were manufactured by the Protein and Peptide Chemistry Facility (PPCF) of the UNIL.
- OVA257-264 and OVA323-339 peptides
- Adjuvants
- Poly (I: C) HMW (tlrl-pic) and Imiquimod R837 (tlrl-imq, InvivoGen)
- CpG-ODN 1826 (Coley Pharmaceuticals. No longer available. CpG-ODN 1826, tlrl-1826 from InvivoGen can be substituted.)
- Quil A saponin mix from Quillaja saponaria (Brenntag Nordic A/S)
- Poly (I: C) HMW (tlrl-pic) and Imiquimod R837 (tlrl-imq, InvivoGen)
- Commercial reagents
- CD45 MACS microbeads, mouse (Miltenyi Biotec, catalog number: 130-052-301 )
Datasheet available online: https://www.miltenyibiotec.com/~/media/Images/Products/Import/0001200/IM0001245.ashx - LIVE/DEAD Aqua cell stain (Life Technologies, InvitrogenTM, catalog number: L34957 )
- CD45 MACS microbeads, mouse (Miltenyi Biotec, catalog number: 130-052-301 )
Equipment
- Falcon™ 40 µm cell strainer (blue) (Corning, catalog number: 352340 )
- 1ml BD Tuberculin Syringe & 26 g or 27 g x 0.5" BD™ PrecisionGlide Needle (BD, catalog number: 3052111 or 305109 )
- Falcon™ 50 ml conical centrifuge tubes (Corning, catalog number: 352070 )
- Falcon™ 15 ml conical centrifuge tubes (Corning, catalog number: 352099 )
- Falcon™ 6 well, non-treated, flat-bottom tissue culture (Corning, catalog number: 351147 )
- Dissection scissors or scalpel blade and fine-tipped forceps or tweezers
- 10 ml BD™ syringe (catalog number: 309604 ) with 18 g or 20 g x 1" BD™ PrecisionGlide needle (BD, catalog number: 305195 or 305175 )
- Haemocytometer
- AutoMACS automatic cell separator (Miltenyi Biotech)
- FACSCanto flow cytometers (BD)
Software
- FlowJo software for Mac, version 9 software (TreeStar)
- Prism Graphpad software
Note: It was used to perform One-way Anova test combined with the Dunnet’s post test for statistical analysis of 3 samples per group, per experiment. Experiments were repeated twice for statistical robustness.
Procedure
- Adoptive cell transfer, tumor challenge and immunization
- Adoptive cell transfers
Antigen-specific CD8+ (OT-I) and CD4+ (OT-II) T cells were isolated from spleens of CD45.2+ T cell receptor (TCR)-transgenic mice. Spleens were disrupted over 40 µm cell strainers with the plastic end of a 1 ml tuberculin syringe plunger and collected into 50 ml Falcon tubes. Total cell numbers were determined by counting with a Haemocytometer. The frequency of OT-I and OT-II cells in the spleens was determined by labelling with TCR Va2 and Vb5.1/5.2 antibodies and analysis by flow cytometry before injection of total splenocytes containing the desired number of antigen-specific T cells (Figure 1). Naive CD45.1 recipient mice received 1e6 OT-I cells and 3e6 OT-II cells in 200 µl of DMEM intravenously in the caudal vein.
Note: T cells can also be transferred by intra-orbital or intra-peritoneal injection if preferred. - Tumor challenge
Mice were challenged the next day with 2e5 B16.OVA tumor cells in 100 µl PBS injected subcutaneously in the left flank. Briefly, mice were restrained using a single-handed hold. About 2/3 of the length of the needle of the tuberculin syringe was inserted into the subcutaneous layer of skin proximal to the joint of the hind leg, at an upward and sideways angle. The correct position of the end of the needle was checked by gently moving the bevel below the skin (just about the hip joint) and confirming it was in loose space. The tumor cells were slowly injected in a single location. The needle was withdrawn carefully and the exit point massaged slightly with the fingertip to prevent escape of the tumor cells. - Immunisations
- One week later, once tumors were palpable, mice were immunised with 10 µg OVA257-264 and 10 µg OVA323-339 peptides in 100 µl PBS subcutaneously at the base of the tail with a 1 ml tuberculin syringe. Briefly, mice were restrained in a holding tunnel with an opening at the end to access the base of the tail. About half the length of the needle of the tuberculin syringe was inserted into the subcutaneous layer of skin just above and to the side of the tail. The correct depth was confirmed by visualizing the bevel just below the skin. The vaccine was slowly injected while keeping pressure at the needle entry point.
Note: A bubble of liquid should become visible under the skin if the correct technique is applied. - Peptides were injected alone or in combination with 50 µg of one of the following adjuvants: CpG-ODN (CpG), HMW Poly (I: C), imiquimod, or Quil A.
- One week later, once tumors were palpable, mice were immunised with 10 µg OVA257-264 and 10 µg OVA323-339 peptides in 100 µl PBS subcutaneously at the base of the tail with a 1 ml tuberculin syringe. Briefly, mice were restrained in a holding tunnel with an opening at the end to access the base of the tail. About half the length of the needle of the tuberculin syringe was inserted into the subcutaneous layer of skin just above and to the side of the tail. The correct depth was confirmed by visualizing the bevel just below the skin. The vaccine was slowly injected while keeping pressure at the needle entry point.
- Adoptive cell transfers
- Tissue harvest and processing
- One week after vaccination, mice were euthanized by CO2 asphyxiation followed by cervical dislocation. Tumors were exposed by cutting through the skin of the flank and excised by carefully grasping with fine-tipped forceps and snipping away the surrounding skin and connective tissue using sharp dissection scissors. Tumors were then placed in pre-weighed 15 ml Falcon tubes containing 2 ml PBS.
- Tubes containing tumors were weighed again and the initial weight of the tube and buffer subtracted to determine tumor mass.
- Tumors larger than 500 µg were cut down to ~500 µg for processing, to avoid overloading of MACS column and as the number of cells obtained in larger tumors are in excess of requirement for flow cytometric analysis.
- Tumors were cleaned of connective tissue and each tumor was placed in a 6 well plate in 1 ml of fresh Tumor digesting buffer. Tumors were dissected by chopping with scissors or by slicing repeatedly with a scalpel blade to produce pieces of ~2 mm2 or less. A further 9 ml of Tumor digesting buffer was added to each well and mixed with the tissue by pipetting gently.
- Tumor tissue was then incubated at 37 °C for one hour to allow digestion, swirling the plate or pipetting the suspension up and down after 30 min to mix.
- At the end of the incubation the tissue pieces were aspirated several times through a 10 ml syringe with a large bore needle (18-20 gauge) to break up remaining tissue and obtain a single cell suspension.
- Cells were washed 2-3 times in 50 ml cDMEM and centrifuged at 1,500 rpm for 10 min to remove excess melanin.
- A rough total cell count was performed using a haemocytometer to determine the amount of MACS beads required.
- One week after vaccination, mice were euthanized by CO2 asphyxiation followed by cervical dislocation. Tumors were exposed by cutting through the skin of the flank and excised by carefully grasping with fine-tipped forceps and snipping away the surrounding skin and connective tissue using sharp dissection scissors. Tumors were then placed in pre-weighed 15 ml Falcon tubes containing 2 ml PBS.
- CD45+ cell selection by magnetic cell sorting with the AutoMACS
- Tumor cells were transferred to 50 ml Falcon tubes and resuspended in 90 µl MACS buffer per 107 cells, as per the manufacturer’s protocol.
- 10 µl anti-CD45 beads were added per 107 cells and the cell and bead mixture was incubated on ice for 15 min with mixing by inversion every 5 min.
- Cells were then washed twice in 50 ml MACS buffer to remove excess beads.
- Cells were then resuspended in 500 µl MACS buffer by gentle pipetting and passed over a 40 µm cell strainer to remove any remaining clumps.
- CD45+ cells were purified by positive selection using magnetic cell separation (MACS) beads and the AutoMACS automatic cell separator. Cell supensions were mixed by vortex at low speed just before passing them through the autoMACS system using a POSSEL program.
- The positive fraction was counted using a haemocytometer to determine the total TIL number before proceeding to FACS staining of the cells.
Notes:- The POSSEL program passes the sample through the autoMACS using a single pass over one magnetic column and is designed to obtain a particular cell population by positive selection from a sample in which a normal to high frequency of the cells express the antigen of interest.
- If an autoMACS machine is not available then manual MACS columns (Miltenyi Biotech) may be used instead. The manual separation procedure can be found in the CD45 MACS microbead datasheet (see section G).
- The purity and yield of the sort can be checked by labeling a small aliquot of the pre-sort and post-sort positive and negative fractions with a CD45 PE antibody and analysis by flow cytometry (Figure 2). After the first 2 experiments, yielding >90% CD45+ cells in the positive fraction, purity testing was no longer performed before the global FACS analysis.
- The POSSEL program passes the sample through the autoMACS using a single pass over one magnetic column and is designed to obtain a particular cell population by positive selection from a sample in which a normal to high frequency of the cells express the antigen of interest.
- Tumor cells were transferred to 50 ml Falcon tubes and resuspended in 90 µl MACS buffer per 107 cells, as per the manufacturer’s protocol.
- Flow cytometry
- ~5 x 106 cells (or the whole sample if less than this number were obtained) were transferred to a 96 well u-bottom plate and washed with PBS.
- Cells were resuspended in 100 µl Live/dead Aqua cell stain at a 1/250 dilution in PBS and incubated for 20 min before washing twice with PBS.
- 25 µl of supernatant from a 2.4G2 (anti-FcgRII monoclonal antibody) producing hybridoma (grown in-house) was added to the samples and incubated on ice for 5-10 min to inhibit nonspecific antibody binding.
- A mix of fluorescent antibodies at 2x final concentration was made in Flow buffer and 25 µl of the mix was added to the cells without first removing the 2.4G2.
- Cells were incubated for a further 20-30 min and the plate was then washed twice with 200 µl per well of Flow buffer.
- Cells were resuspended in 200 µl of Flow buffer and kept on ice until analysis.
- Samples were acquired using LSR-II and FACSCanto flow cytometers.
- Lymphocytes were gated on the basis of forward scatter and side scatter properties and LIVE/DEAD Aqua cell stain was used to exclude dead cells.
- Flow data was analyzed using FlowJo software (Figure 3).
- ~5 x 106 cells (or the whole sample if less than this number were obtained) were transferred to a 96 well u-bottom plate and washed with PBS.
- Multi-colour staining panel
- Live/dead dye Aqua cell stain.
- CD45.1 APC-eFluor780, CD45.2 Pacific Blue, CD8 eFluor700, CD4 PE-Texas Red (optional labeling with Va2 APC and Vb5.1/5.2 PE for more precise identification of TCR transgenic T cells).
- Foxp3 eGFP (endogenous expression).
- As well as the multi-colour staining mix used to label the samples, single stains were made for each fluorochrome (and blood from a Foxp3-eGPF mouse was extracted by nicking of the caudal vein for the GFP single stain).
- The auto compensation option in the BD Diva software was used to compensate the spectral overlap between all the different fluorochromes.
- Live/dead dye Aqua cell stain.
Representative data
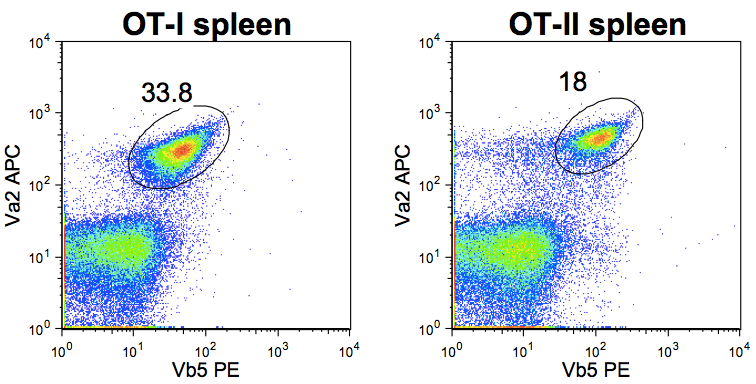
Figure 1. Determining TCR transgenic T cell frequency by Flow cytometry prior to adoptive T cell transfer (lymphocyte population gated based on forward and side scatter properties)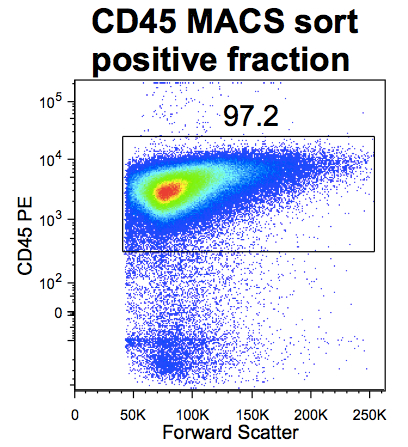
Figure 2. CD45 cell purity after MACS positive selection (single cells first gated based on forward scatter height vs area and live cells detected using Live/dead dye Aqua cell stain)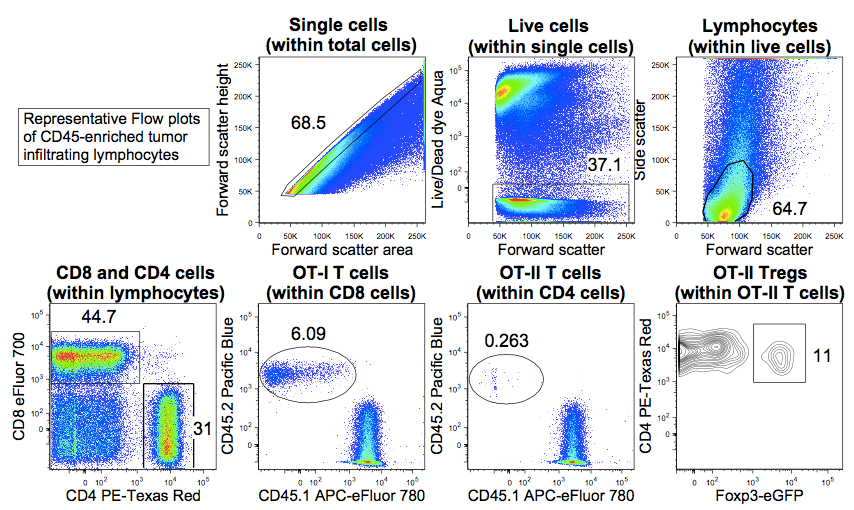
Figure 3. Representative Flow plots showing progressive gating strategy for analysis of tumor infiltrating lymphocytes in CD45 MACS-enriched tumor samples
Recipes
- Complete medium (cDMEM)
DMEM
5% FBS
100 U/ml penicillin & 100 μg/ml streptomycin
25mM Hepes
55 μM 2-ME - Tumor digesting buffer
DMEM
4.4mg/ml Collagenase I
10 µg /ml DNase I - Flow buffer
PBS
2% FBS - MACS buffer
PBS
1% BSA
10 mM EDTA
Acknowledgments
Abbreviated protocol previously published in: Perret et al. (2013). This work was supported by grants from the New Zealand Foundation for Research Science and Technology and the Emma Muschamp Foundation (R. Perret) and from the Swiss National Science Foundation (310030-130812 and CRSII3_141879) and the Medic Foundation (P. Romero). Disclosure of Potential Conflicts of Interest: P. Romero is a consultant/advisory board member of Immatics Biotechnologies, DC Prime, Matwin, and Center for Human Immunology, Pasteur Institute (Paris, France). No potential conflicts of interest were disclosed by the other authors.
References
- Barnden, M. J., Allison, J., Heath, W. R. and Carbone, F. R. (1998). Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol 76(1): 34-40.
- Fidler, I. J. (1975). Biological behavior of malignant melanoma cells correlated to their survival in vivo. Cancer Res 35(1): 218-224.
- Hogquist, K. A., Jameson, S. C., Heath, W. R., Howard, J. L., Bevan, M. J. and Carbone, F. R. (1994). T cell receptor antagonist peptides induce positive selection. Cell 76(1): 17-27.
- Perret, R., Sierro, S. R., Botelho, N. K., Corgnac, S., Donda, A. and Romero, P. (2013). Adjuvants that improve the ratio of antigen-specific effector to regulatory T cells enhance tumor immunity. Cancer Res 73(22): 6597-6608.
- Schuler, P., Contassot, E., Irla, M., Hugues, S., Preynat-Seauve, O., Beermann, F., Donda, A., French, L. E. and Huard, B. (2008). Direct presentation of a melanocyte-associated antigen in peripheral lymph nodes induces cytotoxic CD8+ T cells. Cancer Res 68(20): 8410-8418.
- Wang, Y., Kissenpfennig, A., Mingueneau, M., Richelme, S., Perrin, P., Chevrier, S., Genton, C., Lucas, B., DiSanto, J. P., Acha-Orbea, H., Malissen, B. and Malissen, M. (2008). Th2 lymphoproliferative disorder of LatY136F mutant mice unfolds independently of TCR-MHC engagement and is insensitive to the action of Foxp3+ regulatory T cells. J Immunol 180(3): 1565-1575.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Perret, R., Sierro, S. R., Botelho, N. K., Corgnac, S., Donda, A. and Romero, P. (2014). Analysis of Tumor-infiltrating Lymphocytes Following CD45 Enrichment . Bio-protocol 4(16): e1218. DOI: 10.21769/BioProtoc.1218.
Category
Cancer Biology > General technique > Cell biology assays > Cell isolation and culture
Immunology > Immune cell function > Antigen-specific response
Immunology > Animal model > Mouse
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











