- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Assay for GTP Cyclohydrolase II Activity in Bacterial Extracts
Published: Vol 4, Iss 15, Aug 5, 2014 DOI: 10.21769/BioProtoc.1198 Views: 7560
Reviewed by: Tie LiuFeng LiZhaohui LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

H2 Production from Methyl Viologen–Dependent Hydrogenase Activity Monitored by Gas Chromatography
Nuttavut Kosem
Dec 5, 2023 1769 Views

Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1584 Views

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1927 Views
Abstract
Riboflavin is the precursor of flavin nucleotides FMN and FAD, they play significant roles in all organisms. GTP is the initial precursor on riboflavin biosynthesis pathway and GTP cyclohydrolase II catalyzes the first step of this pathway. It converts GTP to 2,5-diamino-6-ribosylamino-4 (3H) -pyrimidinone 5'-phosphate. This protocol provides a reliable and fast method to assay GTP cyclohydrolase II activity from crude bacterial extracts. The product of the reaction catalyzed by GTP cyclohydrolase II, 2,5-diamino-6-ribosylamino-4 (3H) -pyrimidinone 5'-phosphate, is converted to its fluorescent derivative 6,7-dimethylpterin, which is then separated on a XTerra MS C18 column and detected using fluorescence HPLC system.
Materials and Reagents
- Sinorhizobium meliloti (S. meliloti 1021, Galibert et al., 2001)
Note: The protocol can be also applied to other bacteria.
- Bio-Rad Protein Assay Kit I (Sigma-Aldrich, catalog number: 500-0001 )
- 50 mM Tris-HCl buffer (pH 7.5) (5 ml per sample)
- 1x BugBuster reagent (Novagen, catalog number: 70921 )
- 0.5 M EDTA (pH 8.0) (2.5 µl per sample)
- GTP (Guanosine 5’-triphosphate sodium salt hydrate) (Sigma-Aldrich, catalog number: G8877 )
- HPLC Standard: 6,7-Dimethylpterin (Schircks Laboratories, catalog number: 11.503 )
- Biotin
- YMB medium (Somerville and Kahn, 1983) (see Recipes) (1 plate per sample)
- MMNH4 medium (Somerville and Kahn, 1983) (see Recipes) (13 ml per sample)
- Lysis buffer (see Recipes) (1 ml per sample)
- Desalting buffer (see Recipes) (3 ml per sample)
- RibA assay buffer (see Recipes) (2.5 µl per sample)
- Derivatization reagent (see Recipes) (50 µl per sample)
- HPLC mobile phase (see Recipes)
Note: Except as otherwise noted, all other chemicals were obtained from Sigma-Aldrich.
Equipment
- 14 ml Falcon tubes (BD Biosciences, catalog number: 352059 )
- Eppendorf 1.7 ml tubes (Thermo Fisher Scientific, catalog number: 14-222-168 )
- 0.22 µm syringe filter (Microsolv Technology, catalog number: 58022-N04-C )
- 2 ml Zeba Spin Desalting Columns (Thermo Fisher Scientific, catalog number: 87768 )
- Mini-centrifuge
- Centrifuge
- Shaker
- HPLC: Waters Alliance 2695 HPLC system linked to a 2475 fluorescence detector
- XTerra MS C18 column (4.6 x 100 mm, 5 µm) (Waters, part number: 186000486 )
Procedure
- Cell extract preparation
- Grow S. meliloti on YMB plate for 48 h at 30 °C. The optimal temperature for S. meliloti growth is between 28 °C and 30 °C.
- Inoculate S. meliloti from the stock YMB plate to an OD600 of ~0.1 in 3 ml MMNH4 medium.
- Grow cells for 48 h at 30 °C, 250 rpm in 14 ml Falcon or glass tubes.
- Dilute the cells 20-fold into 10 ml fresh MMNH4 medium and grow overnight at 30 °C, 250 rpm in 50 ml glass tubes or flasks.
- Harvest cells from 10 ml culture by centrifugation at 3,600 x g for 15-20 min at 4 °C in 14 ml Falcon tubes.
- Discard the supernatant and wash the cells with 5 ml 50 mM Tris buffer at 4 °C.
- Resuspend the pellet in 1 ml of lysis buffer at 4 °C.
- Incubate at room temperature for 15 min, without shaking.
- Centrifuge the lysate at maximum speed for 15 min at 4 °C.
- Measure protein concentration in the cell lysates using Bio-Rad Protein Assay Kit.
- Grow S. meliloti on YMB plate for 48 h at 30 °C. The optimal temperature for S. meliloti growth is between 28 °C and 30 °C.
- Enzymatic assay
This assay protocol was adapted from a method for assaying GTP cyclohydrolase II activity in the purified enzyme (Bacher et al., 1997) and modified for using with bacterial extracts.- Desalt the cell lysate using 2 ml Zeba Spin Desalting Columns following the manufacturer’s instructions. The column was equilibrated with 3 ml desalting buffer.
- Add 25 µl of the desalted whole cell lysate form the previous step in a total volume of 50 µl reaction mixture containing 10 µl 5 x RibA assay buffer, 5 µl 10 mM GTP (prepare GTP in ultrapure water).
- Incubate the assay at 37 °C for 30 min.
- Terminate the assay by adding 2.5 µl EDTA (0.5 M, pH 8.0).
- Derivatize the product by adding 50 µl of the derivatization reagent, followed by incubation at 70 °C for 20 min.
- Clear the samples by centrifugation at 3,000 x g for 10 min at 4 °C.
- Filter the supernatant using a 0.22 µm syringe filter.
- Analyze the derivatization products using HPLC with fluorescence detection.
- Desalt the cell lysate using 2 ml Zeba Spin Desalting Columns following the manufacturer’s instructions. The column was equilibrated with 3 ml desalting buffer.
- HPLC purification and signal detection
- Separate the fluorescent products on a XTerra MS C18 column (4.6 x 100 mm, 5 µm) using a Waters Alliance 2695 HPLC system linked to a 2475 fluorescence detector. Excitation and emission wavelengths are 330 nm and 435 nm, respectively.
- Separate the fluorescent products on a XTerra MS C18 column (4.6 x 100 mm, 5 µm) using a Waters Alliance 2695 HPLC system linked to a 2475 fluorescence detector. Excitation and emission wavelengths are 330 nm and 435 nm, respectively.
- Data analysis
- Quantify the products by comparison to standards. A standard curve was obtained by running sequential dilution of 6,7-dimethylpterin (0.25 nM, 2.5 nM, 25 nM, 0.25 µm, 2.5 µM and 25 µM) on HPLC.
- Normalize the enzyme activity against protein concentration and assay incubation time in nmol/min/mg protein.
- Perform three replicates on each assay. Data is the average ± S.E. of three replicates.
Note: The activities of GTP cyclohydrolase II vary in different organisms. The activity of purified recombinant enzymes have been reported from about 0.1- 182 nmol/min/mg protein, the activity in bacterial extracts can be lower (Herz et al., 2000; Kaiser et al., 2002; Yurgel et al., 2014).
- Quantify the products by comparison to standards. A standard curve was obtained by running sequential dilution of 6,7-dimethylpterin (0.25 nM, 2.5 nM, 25 nM, 0.25 µm, 2.5 µM and 25 µM) on HPLC.
Representative data
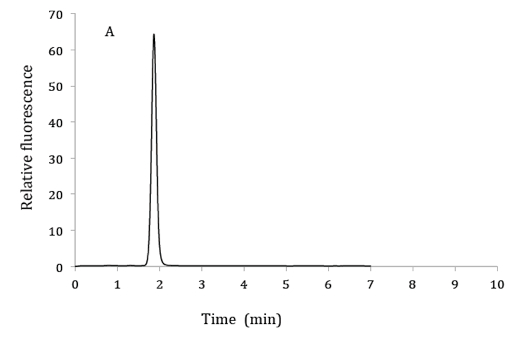
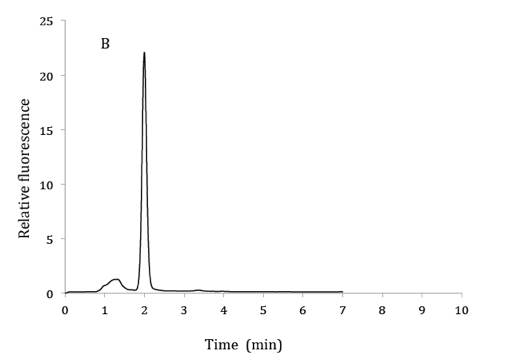
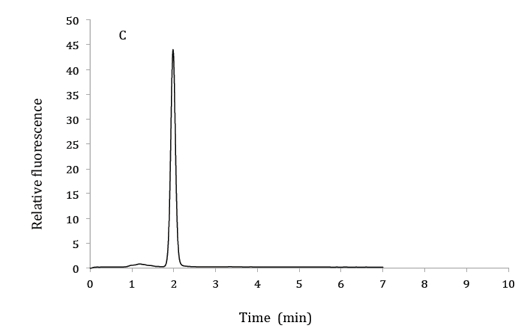
Figure 1. HPLC chromatograms of 2.5 µM 6,7-dimethylpterin standard (A), product of GTP cyclohydrolase II assay from bacterial extracts (B) and co-elution of product from GTP cyclohydrolase II assay with 6,7-dimethylpterin standard (C). HPLC mobile phase: 10% methanol and 90% phosphoric acid (v/v). Excitation and emission wavelengths were 330 nm and 435 nm respectively.
Recipes
- YMB media for Rhizobium
1 L
Concentration
Yeast Extract
1 g
Mannitol
10 g
54.9 mM
Agar
15 g
Autoclave, cool to 55 °C, then add
YMB Salt I
10 ml
YMB Salt II
10 ml
YMB Salt I
1 L
Concentration
K2HPO4
50 g
287.06 mM
NaCl
10 g
171.15 mM
d-H2O
960 ml
YMB Salt II
1 L
Concentration
MgSO4.7H2O
20 g
81.11 mM
d-H2O
1 L
- MMNH4 (minimal mannitol ammonia media for Rhizobium)
per 1 L
Concentration
Mannitol
10.0 g
54.9 mM
NH4Cl
0.5 g
9.34 mM
Agar (for plates preparation)
15.0 g
d-H2O
970 ml
Autoclave, cool to 55 °C, then add:
Biotin (0.2 mg/ml in 50% EtOH)
1.0 ml
Thiamine (2 mg/ml), filter sterilized
1.0 ml
Min Man Salts I
10.0 ml
Min Man Salts II
10.0 ml
Min Man Salts I
per 1 L
Concentration
K2HPO4
100 g
574.12 mM
KH2PO4
100 g
734.8 mM
Na2SO4
25 g
174.8 mM
d-H2O
1 L
Min Man Salt II
per 1 L
Concentration
FeCl3.6H2O
1.0 g
3.7 mM
Concentrated HCl
adjust pH to ~7.0 (~1drop)
CaCl2.2H2O
10.0 g
68 mM
MgCl2.6H2O
25.0 g
123 mM
d-H2O
1 L
Autoclave
- Lysis buffer
50 mM Tris-HCl (pH 7.5)
10 mM MgCl2
1 mM Tris (hydroxypropyl) phosphine
1x BugBuster reagent
- Desalting buffer
50 mM Tris-HCl (pH 7.5)
1 mM Tris (hydroxypropyl) phosphine
10% glycerol
- RibA assay buffer
100 mM Tris-HCl (pH 8.5) buffer
5 mM MgCl2
5 mM dithiothreitol (DTT)
- Derivatization reagent
1% (v/v) diacetyl
15% (w/v) trichloroacetic acid
- HPLC mobile phase
10% (v/v) methanol
90% phosphoric acid (27 mM phosphoric acid)
Acknowledgments
This protocol was adapted from Yurgel et al., 2014. This work was supported by the Agricultural Research Center (WNP-00773) at Washington State University and grant DE-FG0396ER20225 Vol. 27, No. 5, 2014 / 445 from the Energy Biosciences Program at the United States Department of Energy, and by grant NSF-MCB 1052492 to S. Rajamani. This activity was funded, in part, with an Emerging Research Issues Internal Competitive grant from the Agricultural Research Center at Washington State University, College of Agricultural, Human, and Natural Resource Sciences and Biologically-Intensive Agriculture and Organic Farming (BIOAg) Internal Competitive grant from The Center for Sustaining Agriculture and Natural Resources (CSANR) at Washington State University to S. Yurgel. We thank M. Kahn for discussions and the Washington State University Laboratory for Biotechnology and Bioanalysis for sequencing support.
References
- Bacher, A., Richter, G., Ritz, H., Eberhardt, S., Fischer, M. and Krieger, C. (1997). Biosynthesis of riboflavin: GTP cyclohydrolase II, deaminase, and reductase. Methods Enzymol 280: 382-389.
- Galibert, F., Finan, T. M., Long, S. R., Puhler, A., Abola, P., Ampe, F., Barloy-Hubler, F., Barnett, M. J., Becker, A., Boistard, P., Bothe, G., Boutry, M., Bowser, L., Buhrmester, J., Cadieu, E., Capela, D., Chain, P., Cowie, A., Davis, R. W., Dreano, S., Federspiel, N. A., Fisher, R. F., Gloux, S., Godrie, T., Goffeau, A., Golding, B., Gouzy, J., Gurjal, M., Hernandez-Lucas, I., Hong, A., Huizar, L., Hyman, R. W., Jones, T., Kahn, D., Kahn, M. L., Kalman, S., Keating, D. H., Kiss, E., Komp, C., Lelaure, V., Masuy, D., Palm, C., Peck, M. C., Pohl, T. M., Portetelle, D., Purnelle, B., Ramsperger, U., Surzycki, R., Thebault, P., Vandenbol, M., Vorholter, F. J., Weidner, S., Wells, D. H., Wong, K., Yeh, K. C. and Batut, J. (2001). The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293(5530): 668-672.
- Herz, S., Eberhardt, S. and Bacher, A. (2000). Biosynthesis of riboflavin in plants. The ribA gene of Arabidopsis thaliana specifies a bifunctional GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate synthase. Phytochemistry 53(7): 723-731.
- Kaiser, J., Schramek, N., Eberhardt, S., Puttmer, S., Schuster, M. and Bacher, A. (2002). Biosynthesis of vitamin B2. Eur J Biochem 269(21): 5264-5270.
- Somerville, J. E. and Kahn, M. L. (1983). Cloning of the glutamine synthetase I gene from Rhizobium meliloti. J Bacteriol 156(1): 168-176.
- Yurgel, S. N., Rice, J., Domreis, E., Lynch, J., Sa, N., Qamar, Z., Rajamani, S., Gao, M., Roje, S. and Bauer, W. D. (2014). Sinorhizobium meliloti flavin secretion and bacteria-host interaction: role of the bifunctional RibBA protein. Mol Plant Microbe Interact 27(5): 437-445.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yurgel, S. N., Sa, N., Rice, J. and Roje, S. (2014). Assay for GTP Cyclohydrolase II Activity in Bacterial Extracts. Bio-protocol 4(15): e1198. DOI: 10.21769/BioProtoc.1198.
Category
Microbiology > Microbe-host interactions > In vivo model > Plant
Microbiology > Microbial biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









