- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Novel Method for Site-specific Induction of Oxidative DNA Damage to Study Recruitment of Repair Proteins to Heterochromatin and Euchromatin
Published: Vol 4, Iss 11, Jun 5, 2014 DOI: 10.21769/BioProtoc.1140 Views: 10327
Reviewed by: Fanglian HeClaudia CatalanottiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protocol for Quantifying γH2AX Foci in Irradiated Cells Using Immunofluorescence and Fiji Software
Lu Deng [...] Lingying Wu
Aug 20, 2025 2448 Views

Colocalizing Telomeres With PML or γH2AX Foci by IF-FISH in Mouse Brain Neurons
Anna Konopka
Nov 5, 2025 1515 Views

A Quantitative DNA Fiber Assay to Monitor Replication Fork Progression, Protection, and Restart
Debanjali Bhattacharya and Ganesh Nagaraju
Feb 5, 2026 225 Views
Abstract
ROS-induced DNA damage is repaired in living cells within a temporal and spatial context, and chromatin structure is critical to a consideration of DNA repair processes in situ. It’s well known that chromatin remodeling factors participate in many DNA damage repair pathways, indicating the importance of chromatin remodeling in facilitating DNA damage repair. To date, there has been no method to induce site-specific oxidative DNA damage in living cells. Therefore, it is not known whether the DNA repair mechanisms differ within active or condensed chromatin. We recently established a novel method, DTG (Damage Targeted at one Genome-site), to study DNA damage response of reactive oxygen species (ROS)-induced DNA damage in living cell at one genome loci with active or inactive transcription. For this, we integrated a tetracycline responsive elements (TRE) cassette (~90 kb) at X-chromosome in U2OS cells (Lan et al., 2010), then fused KillerRed (KR), a light-stimulated ROS-inducer which can specifically produce ROS-induced DNA damage, to a tet-repressor (tetR-KR, OFF) or a transcription activator (TA-KR, ON) (Lan et al., 2014) (Figure 1). TetR-KR or TA-KR binds to the TRE cassette and induces ROS damage under hetero- or euchromatin states, respectively. How chromatin states regulate the DNA damage response processes can be examined by using this powerful method.
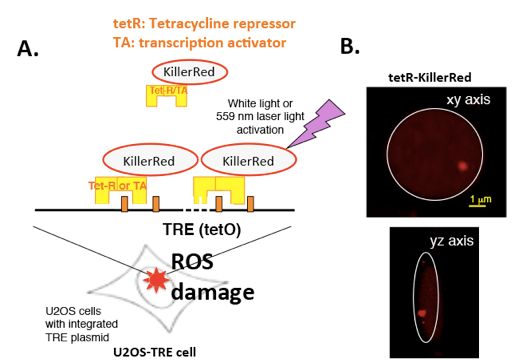
Figure 1. Scheme of the DTG system. A. Scheme of tetR and TA tagged KR expression in the U2OS TRE cell line. To induce ROS-mediated damage at a specific locus in the genome, we fused KR to the tetracycline repressor to induce ROS damage in a 90 kb tetracycline response element (TRE) array (totally 96 repeats) in U2OS cells. B. Expression of tetR-KR in U2OS TRE cell line.
Materials and Reagents
- U2OS-SCE cell line (made in our laboratory) (Lan et al., 2010)
Notes:- In this cell line, 200 copies of pTRE / I-SceI were integrated in U2OS cells.
- Cells are cultured with Dulbecco’s Modified Eagle’s Medium High glucose with stable L-glutamine (DMEM) (EuroClone S.p.A. P.IVA, catalog number: ECM0103L ) with 10% Fetal Bovine Serum (FBS) (Sigma-Aldrich, catalog number: F9665 ).
- For preparation of cells, cells were washed with PBS without Ca2+ and Mg2+ (EuroClone S.p.A. P.IVA, catalog number: ECB4004L ), trypsinized with Trypsin/EDTA without out Ca2+ and Mg2+ (Thermo Fisher Scientific, catalog number: BW17161E ).
- In this cell line, 200 copies of pTRE / I-SceI were integrated in U2OS cells.
- Plasmids
- pBROAD3/tetR-KR
- pBROAD3/tetR-mCherry
- pBROAD3/TA-KR
- pBROAD3/TA-mCherry
Note: pBROAD3/tetR-mcherry was provided by Dr. Edith Heard (Masui et al., 2011); pBROAD3/tetR-KR, pBROAD3/TA-mCherry, pBROAD3/TA-KR were made in our laboratory (Lan et al., 2014). Sequences are available based on requests.
- pBROAD3/tetR-KR
- GFP-fusion protein or endogenous protein
- Lipofectamine 2000 (Life Technologies, catalog number: 12566-014 )
- Opti-MEM (Life Technologies, catalog number: 51985-091 )
- 35 mm glass bottom culture dishes (MatTek, catalog number: P35GC-1.5-14-C )
- IMMOIL-F30CC (Chip Humphries, catalog number: Z-81225 )
Equipment
- 35 mm glass bottom culture dishes
- 37 °C, 5% CO2 cell culture incubator
- Olympus FV1000 confocal microscopy system (OLYMPUS, model: FV1000- FILTER DETECT ; SYS.PACKAGE: IX81-1 405/M_AR/559/635N) with 488, 559 nm lasers
- PLAPON 60x oil lens (super chromatic abe. corr. obj W/1.4NA FV) (OLYMPUS, catalog number: FM1-U2B990 )
- Thermo-plate (MATS-U52RA26 for IX81/71/51/70/50, metal insert, HQ control) (OLYMPUS, catalog number: OTH-I0126 )
- 15 watt cool white fluorescent bulb (OSRAM SYLVANIA)
- A stage UVP (Upland, CA)
Procedure
- Seed U2OS-SCE cells into 35 mm glass bottom culture dishes at an approximate density of 5 x 105 per dish in DMEM with 10% FBS at 37 °C incubator with 5% CO2 for overnight.
- For GFP-fusion protein, co-transfect 1 µg GFP-Fusion protein with any 1 µg of pBROAD3/tetR-KR, pBROAD3/tetR-mCherry, pBROAD3/TA-KR and pBROAD3/TA-mCherry using Lipofectamine 2000. For endogenous protein, just transfect anyone of pBROAD3/tetR-KR, pBROAD3/tetR-mCherry, pBROAD3/TA-KR and pBROAD3/TA-mCherry. Follow the transfection protocol of Lipofectamine 2000.
- Then incubate in the 5% CO2 incubator at 37 °C for 24 h to 48 h.
- KR activation (There are two ways for KR activation.)
- Local activation of one KR spot
Local activation of one KR spot was performed with a 559 nm laser (1 mW/scan) of Olympus FV1000 confocal microscopy system in a selected area. One scan takes less than 1 sec. The final power (160 mJ) delivered to the KR (around 1 µm2) spot is around 6 mJ/µm2.
- Activation of KR in bulk cells
Activation of KR in bulk cells was done by exposing cells to a 15 watt SYLVANIA cool white fluorescent bulb for 10 min in a stage UVP.
- Local activation of one KR spot
- The response of either GFP-tagged protein or endogenous protein of authors at the sites of tetR or TA-KR can be monitored in living cell.
- The response of endogenous protein at the sites of tetR or TA-KR can be monitored by immunostaining with the antibody.
Notes
For calculation of the dose that was delivered to the KillerRed spot:
- In the case of the 559 nm laser, the laser light was delivered to the selected area (around 25 m2) with 20 mJ/sec for 8 sec. Therefore, the final power (160 mJ) delivered to the KR (around 1 µm2) spot is around 6 mJ/µm2. For calculation of the dose that was delivered to the KillerRed spot based on the pixel size, the pixel size for irradiation is (0.138 µm/pixel) and the dwell time per pixel is (8 us/pixel). The irradiation is at 1.0 mW (1.0 mJ/s). With a dwell time of 8 us/pixel, this irradiates each pixel with 8.0 nJ/pixel/scan. Multiplying by the number of scans gives the total energy per pixel.
- In the case of fluorescent light activation, the rate of light is 15 J/m2/sec. With a 10 min light exposure, 9,000 J were delivered to the whole dish; the final power delivered to the KR (around 1 µm2) spot is around 9 mJ/µm2 upon light exposure. Cells were placed under a water bottle (height to light is 15 cm) to prevent an increase of temperature.
Acknowledgments
This protocol has been adapted from Lan et al. (2010) and Lan et al. (2014).
References
- Lan, L., Nakajima, S., Wei, L., Sun, L., Hsieh, C. L., Sobol, R. W., Bruchez, M., Van Houten, B., Yasui, A. and Levine, A. S. (2014). Novel method for site-specific induction of oxidative DNA damage reveals differences in recruitment of repair proteins to heterochromatin and euchromatin. Nucleic Acids Res 42(4): 2330-2345.
- Lan, L., Ui, A., Nakajima, S., Hatakeyama, K., Hoshi, M., Watanabe, R., Janicki, S. M., Ogiwara, H., Kohno, T. and Kanno, S.-i. (2010). The ACF1 complex is required for DNA double-strand break repair in human cells. Mol Cell 40(6): 976-987.
- Masui, O., Bonnet, I., Le Baccon, P., Brito, I., Pollex, T., Murphy, N., Hupe, P., Barillot, E., Belmont, A. S. and Heard, E. (2011). Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell 145(3): 447-458.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wei, L., Nakajima, S., Levine, A. S. and Lan, L. (2014). Novel Method for Site-specific Induction of Oxidative DNA Damage to Study Recruitment of Repair Proteins to Heterochromatin and Euchromatin. Bio-protocol 4(11): e1140. DOI: 10.21769/BioProtoc.1140.
Category
Cell Biology > Cell structure > Chromosome
Molecular Biology > DNA > DNA damage and repair
Biochemistry > Other compound > Reactive oxygen species
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










