- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Microfluidic-based Time-kill Kinetic Assay
Published: Vol 4, Iss 9, May 5, 2014 DOI: 10.21769/BioProtoc.1116 Views: 12769
Reviewed by: Fanglian HeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Rapid Detection of Proliferative Bacteria by Electrical Stimulation
Conor LA Edwards [...] Munehiro Asally
Feb 5, 2020 6566 Views

Tracking the Subcellular Localization of Surface Proteins in Staphylococcus aureus by Immunofluorescence Microscopy
Salvatore J. Scaffidi [...] Wenqi Yu
May 20, 2021 7813 Views

Multiplexed Microfluidic Platform for Parallel Bacterial Chemotaxis Assays
Michael R. Stehnach [...] Jeffrey S. Guasto
Sep 5, 2024 2055 Views
Abstract
In many environments, bacteria favor a sessile, surface-attached community lifestyle. These communities, termed biofilms, are ubiquitous among many species of bacteria. In some cases, biofilms form under flow conditions. Flow chambers, and in particular microfluidic channels, can be used to observe biofilm development and physiological effects while varying nutrient conditions, flow velocities, or introducing antimicrobials to the biofilm in real time. Here, we describe a microfluidic-based kill-kinetics assay for the observation of antimicrobial effects on biofilms under flowing conditions.
Materials and Reagents
- Glycerol stock of Pseudomonas aeruginosa (P. aeruginosa) (acquired from a single colony, stored at -80 °C)
- Bacto-tryptone (BD Biosciences, catalog number: 211705 )
- Sodium chloride (Sigma-Aldrich, catalog number: S7653 )
- Colistin sodium sulfate (Sigma-Aldrich, catalog number: C4461 )
- Polydimethylsiloxane (PDMS) (Sylgard 184 Silicone Elastomer Kit) (Dow Corning Corporation)
- Petri dishes (150 mm x 15 mm) (VWR International, catalog number: 25384-326 )
- 100% Ethanol
- Sterile dH2O
- LIVE/DEAD BacLight Bacterial Viability Kit with Syto9/Propidium Iodide (Life Technologies, catalog number: L-7012 )
- 1% TB liquid medium (see Recipes)
Equipment
- Sterile 14 ml polystyrene culture tubes (VWR International, catalog number: 60818-725 )
- Sterile 1 μl plastic loops (Santa Cruz, catalog number: SC-200266 )
- Spectrophotometer for absorbance readings
- 37 °C shaking incubator
- Microfluidic channel molds (Figure 1)
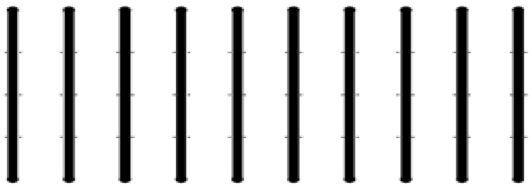
Figure 1. Microfluidic channels. Each channel is 100 μm deep and 500 μm wide. The 10 channels were fabricated by depositing SU-8 2150 on silicon wafers using standard soft lithography techniques (Xia and Whitesides, 1998). This device allows for 10 parallel experiments. - SU-8 2150 (MicroChem Corp.)
- Glass Slides (75 x 50 mm) (Ted Pella, catalog number: 26005 ) or cover glass (75 x 50 mm) (Ted Pella, catalog number: 260462 )
- Lens paper
- Desiccator with vacuum line
- Exacto-knife or scalpel
- 5 ml syringes (BD, catalog number: 309647 )
- Polyethylene tubing (ID 0.58 mm, OD 0.965) (BD, IntramedicTM, Clay Adams®, catalog number: 427411 )
- Harris Unicore hole punch (1.20 mm) (Ted Pella, catalog number: 15074 )
- Hand-held corona generator (Laboratory Corona Treater, Electro-Technic Products, Model: BD-20AC )
- Syringe pump with 10-syringe holder adaptor (Harvard Apparatus)
- Fluorescence microscope with automated focus/stage system (e.g., Nikon TE2000-E equipped with an Andor iXon-885 and a 40x long working distance objective)
Software
- ImageJ
Procedure
- Microchannel molds (Figure 1) were prepared by depositing SU-8 2150 on silicon wafers using standard soft lithography techniques (Xia and Whitesides, 1998).
Note: Stanford Microfluidics Foundry (http://www.stanford.edu/group/foundry/index.html) or the SIMTech Microfluidics Foundry in Singapore (http://www.simtech.a-star.edu.sg/smf/) provide services for fabrication. The mask pattern is included as a pdf supplemental file. - Microfluidic device assembly:
- Place the mold into a 150 mm x 15 mm petri dish with the patterned side facing up.
- Prepare 25 ml of the PDMS mixture. In a plastic weigh-boat, weigh 10 parts silicon elastomer then add 1 part curing agent. Mix thoroughly for at least one minute, and then pour over the silicon master.
- Place the dish in a desiccator and degas under vacuum for 1 h. It is important to remove all air bubbles from the PDMS before curing.
- Cure the PDMS for 24 h in a 37 °C incubator, or, alternatively, for 2 h at 65 °C.
Note: Read the manufacturer’s suggestions for curing PDMS. - The cured PDMS should be firm to the touch. Using a sharp blade (Exacto-knife or scalpel) carefully cut out all of the lanes in a single, rectangular piece. The patterned microchannels will be visible in the PDMS. Cover channels with cellophane tape to protect from dust particles and debris.
- Punch holes at the channel inlets and outlets with the 1.2 mm Harris Unicore hole punch. Take care that the PDMS plugs are removed. Remove cellophane tape. Thoroughly clean the PDMS device and a glass slide with 100% ethanol and lens paper to remove any debris.
- Place the glass slide and PDMS channel side up on a clean flat surface. Hold the hand-held corona generator approximately 1-2 cm above the surface. Treat for 30 sec uniformly over each surface.
- Carefully place the PDMS device on the treated glass slide, with the microchannel pattern facing the glass slide. Apply gentle, uniform pressure by hand to ensure a uniform seal. Care must be taken not to apply too much pressure, which can lead to channel collapse. After a final bonding step at 65 °C for 15 min, the device is ready to use.
- Place the mold into a 150 mm x 15 mm petri dish with the patterned side facing up.
- Prepare up to five 14 ml polystyrene culture tubes with 5 ml of 1% TB. Inoculate by carefully scraping the surface of the bacterial strain glycerol stock. For this device, 5 separate strains can be tested using the 10 microchannels. For each strain, one microchannel will serve as a control and the other will be used for an antimicrobial challenge.
- Place the cultures in a 37 °C incubator, shaking at 250 rpm overnight.
- Remove the culture from the incubator and prepare another 14 ml polystyrene culture tube with 5 ml of 1% TB. Inoculate the culture with 50 μl from the overnight culture.
- Incubate in the 37 °C incubator, shaking at 250 rpm for approximately 4 h, monitoring until an OD600 of 0.5 is reached indicating mid-exponential phase. Use this time to sterilize the device.
- Dilute the culture to an OD600 of 0.0025 in 5 ml of 1% TB.
- Insert tubing in the inputs and outputs of each microchannel, applying enough pressure to force the tubing roughly half-way from the PDMS surface to the glass slide but carefully to avoid cracks in the PDMS inlet and outlets.
- Attach the output tubing to a 5 ml syringe equipped with a BD 23G1 gauge needle for each microchannel. Place the syringes on to the syringe pump and mount the device onto the microscope stage (Figure 2). Set the diameter of the syringe on the syringe pump.
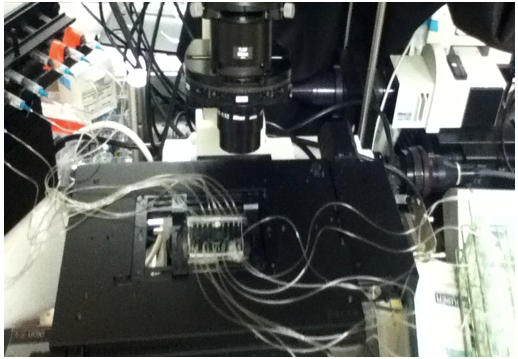
Figure 2. Microfluidic device setup. The microfluidic device is mounted onto the microscope stage for the duration of the experiment. A syringe pump equipped with a 10-syringe holder withdraws fluid from the culture tube reservoirs, flowing it through each of the 10 channels run in parallel. - Place the input tubing into a reservoir of 100% ethanol. Set the syringe pump to withdrawal. Sterilize the tubing and microchannels by flowing 100% ethanol into the channels for a minimum of 15 min and rinse with sterilized water.
Note: Fluid is replaced in the reservoir by adding a separate line of tubing attached to a free syringe into the bottom of the reservoir. After stopping the flow, the fluid is manually removed and replaced by pipetting in new volume of fluid. This step facilitates exchange between fluids without introduction of air bubbles. - Once the device is sterilized and rinsed, place the input tubing into each of the prepared cultures and start the syringe pump to withdraw the medium from the cultures at a flow rate of 25 μl/min. Withdraw at this rate for 15 min or until the culture medium has reached the microchannels.
- Set the flow rate to 0.5 μl/min for 18 h. This will facilitate attachment of P. aeruginosa, yielding a monolayer of cells.
Note: For longer time points, a more mature, thicker biofilm requires a confocal microscope for 3 dimensional quantification via z-stack reconstruction. - After 18 h, replace the culture medium in the reservoir tubes with 3 ml of 1% TB. Wash each microchannel at a flow rate of 25 μl/min. Using the microscope stage/focus positioning software, program 3 fields of view for each microchannel. Include image capture every 5 min for 2 h using phase contrast and filter cubes corresponding to the excitation/emission wavelengths of Syto 9 and Propidium Iodide.
- Prepare the Syto 9/Propidium Iodide viability dye at a 1:1 ratio. Mix 6 μl of dye solution for 1 ml of dH2O to achieve the final working concentration of dye.
Note: After attempting several solvents, dH2O proved to be the most efficient. Other solvents such as 1% TB medium, 1x PBS, and nanopure H2O resulted in decreased binding of both dyes. - Introduce the dye into the microchannel at a flow rate of 25 μl/min for approximately 15 min. Stop flow. Let the dye remain in the channel for 15-30 min.
Note: For the next step, it is important to know when the antibiotic solution reaches the microchannel since time zero is a critical data point for the kill kinetics curve. This is simple to calculate by monitoring the time that it takes for the dye to reach the microchannel at the suggested flow rate. Make note of this time for the next step in the protocol. - Prepare a solution of 20 μg/ml of colistin in 1% TB. Remove residual dye from the reservoir tubes for the microchannel inputs. For each strain, add 1 ml of the colistin solution to the first microchannel and 1 ml of dH2O to the second microchannel to serve as a control. Initiate flow at 25 μl/min and reduce flow rate to 0.5 μl/min once the antibiotic has reached the microchannel. Start the imaging software at time zero (point when the antibiotic reaches the microchannels) to record images at the designated fields of view.
- As cell death progresses, an increase in red cells will be observed for antibiotic susceptible cells and a decrease in green cells as propidium iodide quenches the Syto 9.
- The total number of dead cells in the biofilms for each time point can be quantified in ImageJ (Schneider et al., 2012) by converting the fluorescence image to an 8-bit binary image. Apply a constant threshold to each image to subtract background fluorescence. The final data can be expressed as a percentage of the total biofilm area (phase-contrast images) for each time point.
Recipes
- 1% TB liquid medium (1 L)
10 g of Bacto-tryptone
2.5 g Sodium chloride
Add dH2O to 1 L
Sterilize by autoclaving on liquid cycle
Acknowledgments
The development of this protocol was funded by the following: NIH Grant R01 EB017755, NIH-NIEHS Training Grant in Toxicology T32 ES7020-37, NSF OCE-0744641-CAREER, NIH 1R01GM100473, NSF CBET-0966000.
References
- Billings, N., Millan, M., Caldara, M., Rusconi, R., Tarasova, Y., Stocker, R. and Ribbeck, K. (2013). The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog 9(8): e1003526.
- Schneider, C. A., Rasband, W. S. and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7): 671-675.
- Xia, Y. and Whitesides G. M. (1998). Soft lithography. Angewandte Chemie-International Edition 37(5): 551-575.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Billings, N., Rusconi, R., Stocker, R. and Ribbeck, K. (2014). Microfluidic-based Time-kill Kinetic Assay. Bio-protocol 4(9): e1116. DOI: 10.21769/BioProtoc.1116.
- Billings, N., Millan, M., Caldara, M., Rusconi, R., Tarasova, Y., Stocker, R. and Ribbeck, K. (2013). The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog 9(8): e1003526.
Category
Microbiology > Microbial biofilm > Killing assay
Microbiology > Microbial cell biology > Cell imaging
Cell Biology > Cell imaging > Microfluidics
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









