- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protocol for the Preparation of Arabidopsis Meiotic Chromosome Spreads and Fluorescent in situ Hybridization
Published: Vol 4, Iss 8, Apr 20, 2014 DOI: 10.21769/BioProtoc.1102 Views: 16232
Reviewed by: Renate Weizbauer

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2322 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views
Abstract
This protocol is a more detailed version of previous protocols (Yang et al., 2011; Bolaños-Villegas et al., 2013) developed for the examination of meiotic chromosome spreads. Meiotic chromosome spreads are useful to determine the presence of defects in chromosome pairing and segregation. The protocol also describes how to perform fluorescent in situ hybridization experiments with a centromere probe used to label chromosomes.
Keywords: MeiocyteMaterials and Reagents
- Inflorescences from young, healthy Arabidopsis thaliana plants
- Agar plates, with 1/2x Murashige and Skoog solid medium
- Pectolyase (Sigma-Aldrich, catalog number: P3026 )
- Cellulase (Sigma-Aldrich, catalog number: C1184 )
- β-glucoronidase (Roche Diagnostics, catalog number: 03707598001 )
- Sucrose (Affymetrix, catalog number: 21938 )
- Ethanol (EMD Millipore, catalog number: 1085430250 )
- Chloroform (EMD Millipore, catalog number: 102442 )
- Glacial acetic acid (EMD Millipore, catalog number: 100063 )
- Citric acid anhydrous (Affymetrix, catalog number: 13729 )
- Sodium citrate dihydrate (Affymetrix, catalog number: 13735 )
- Formamide (Sigma-Aldrich, catalog number: F9037 )
- Saline-sodium citrate (SSC) (Sigma-Aldrich, catalog number: S0902 )
- Dextran sulfate (Sigma-Aldrich, catalog number: 42867 )
- Salmon sperm (Life Technologies, catalog number: 15632-011 )
- VECTASHIELD® mounting medium with DAPI (10 ml) (Vector Laboratories, catalog number: H-1400 )
- Fluorescein High-Prime labeling kit (Roche Diagnostics, catalog number: 11585622910 )
- pAL1 centromere probe (ABRC, catalog number: CD3-16 )
- Taq DNA polymerase with standard Taq buffer (New England Biolabs, catalog number: M0273S )
- QIAquick Gel Extraction Kit (kit for purification of DNA from gels) (QIAGEN, catalog number: 28704 )
- Gel Pilot 1 Kb Ladder (molecular weight marker for DNA) (QIAGEN, catalog number: 239085 )
- Agarose for electrophoresis (Bio-Rad Laboratories, catalog number: BR 161-3100 )
- Carnoy’s solution (see Recipes)
- 10 mM Sodium citrate buffer (see Recipes)
- Enzyme digestion buffer (see Recipes)
- FISH buffer (see Recipes)
- 10x PBS (see Recipes)
- 20x SSC (see Recipes)
Equipment
- Hypodermic needles and syringe (Terumo Medical Corporation, model: 26G x ½”, catalog number: NN-2613R )
- Tweezers (Dumont)
- Poly-Prep® poly-L-lysine coated slides (Sigma-Aldrich, catalog number: P0425-72EA )
- Nunc® Lab-Tek® II slides (Thermo Fisher Scientific, catalog number: 154453 )
- Coverslips (Matsunami Glass, catalog number: C218181 )
- Hybridisation oven OV1 (Biometra, catalog number: 052-090 )
- Thermal cycler (Biometra, catalog number: 050-551 )
- Dry block (Thermo Fisher Scientific, Reacti-Therm®)
- -86 °C refrigerator (Thermo Fisher Scientific, model: Forma® 88000 )
- Frigidaire -20 °C refrigerator (Frigidaire, model: FCFS201LFB )
- Fluorescence microscope
- Coplin staining jars (Thermo Fisher Scientific, catalog number: 107 )
- Rubbermaid LunchBlox® food containers
- Super Pap Pen’ liquid blocker pen (Electron Microscopy Sciences, catalog number: 71310 ) (optional)
Procedure
- Preparation of Arabidopsis meiotic chromosome spreads
Note: The most important aspect of this experiment is to collect flower buds at the right stage to isolate anthers that contain male meiocytes. Typically flower buds come in racemes, and only 3-4 are usable. Their size cannot exceed 1/4 of the length of the mouth of a Terumo® 26G x ½” hypodermic needle (approx.1 mm). Anthers should be green. In order to prepare a single slide 5-10 flower buds are required.
- Prior to anther collection and fixation remove the enzyme digestion solution from the refrigerator and allow it to thaw on ice. In the meantime set a hybridization oven at 37 °C, and prepare a lunch box in which the bottom is covered by moist tissues. On the bottom of the lunch box place a rack for PCR tubes, and place the slides on top to avoid direct contact with the wet tissues. Digestion of the anthers allows free meiocytes to be released onto slides where they can be stained and observed under the microscope.
- Racemes are placed first on 1/2x Murashige and Skoog agar plates. Flower buds are excised with tweezers and then placed in a drop of double distilled water (20 μl) in a Lab-Tek® II slide. Lab-Tek® II slides are perfect for this experiment since their coating keeps water drops in place, something regular poly-L-lysine slides cannot do.
- To remove water or any other solution, press the mouth of the needle against the slide, and lift the piston of the syringe slowly (the piston can be lifted slightly before placing the syringe against the slide). Choose an area away from the buds. If possible use the tweezers to stretch the droplet away from the buds.
- Once all flower buds are placed on water, work can begin. Individual flower buds are moved onto a dry area within the slide. After 10 sec the buds mostly dry out and can be dissected at ease. For dissection two tools are used: a tweezer and two hypodermic needles (26G x ½”). Flower buds are placed horizontally and cut at the base with the needles. This allows for quick removal of the petals. Once petals are removed the anthers can be moved individually into a drop of water with the aid of the needles. It is important to maintain the needles dry to allow easy manipulation of the remaining anthers. Yellow anthers are not meiotic anthers and should not be used at all. Anthers must remain moist at all times.
- Once anthers are all placed together, water is removed, leaving only 5 μl. In this way all buds can be further aggregated with the needle. Then add Carnoy’s fixative 20 μl for 20 min, and replenish every 5 min to avoid evaporation (as an option the Super Pap Pen’ liquid blocker pen can be used to better contain the Carnoy’s fixative). Carnoy´s fixative contains no formaldehyde, gluteraldehyde or chloroform, therefore it has little toxicity and allows subsequent ease of manipulation. After 20 min have the fixative removed but leave at least 5 μl. Aggregate the anthers. Then quickly add water, and have it quickly removed (but never totally, leave 5 μl). Then change water two more times (5 min each).
- Add citrate buffer (20 μl). This buffer is used to prepare anthers for enzymatic digestion.
- After removal of citrate buffer add 20-40 μl of enzyme solution to the slide, place a Lab-Tek® II plastic cap on top of the slide (each slide comes with a cap), place the slides inside the lunch box, close it well, and have it placed inside the hybridization oven. Allow digestion of the buds for 2 h at 37 °C. Once digestion is finished, remove the slides, and remove most of the solution (leave 5 μl). Then quickly add 20 μl of citrate buffer, and have it quickly removed (never totally, leave 5 μl), then change the citrate buffer two more times (5 min each) and leave only 10 μl on the slide. Remove the citrate buffer slowly and carefully. If citrate is removed too quickly most anthers will be removed as well and the slides may contain few or no meiotic spreads.
- At this point use the needle, or the tweezers to squeeze the anthers. This allows the release of free meiocytes onto the slide. Do it slowly, for about 10 min. It is important to squeeze slowly and carefully; otherwise the anthers will adhere to each other. At this point the solution will turn turbid, which is normal. Add buffer if needed.
- Add 20 μl of 60% acetic acid, and mix slowly. The solution will turn transparent. Do not allow to evaporate. Acetic acid causes meiocytes to swell, thereby improving the quality of the final chromosome spreads. Transfer 5 μl of the resulting cells suspension to four different poly-L-lysine slides. Place a glass coverslip on top, but before placing it carefully misalign the coverslip by 45 degrees to either side to allow protrusion of one corner. Fold a piece of paper tissue and place a slide inside, then press firmly from above with your thumb. It is very important to keep your thumb in place to prevent displacement of the coverslip. Application of pressure allows bursting of the cells and removal of cytoplasm, while the use of paper tissue prevents the slide from breaking while applying pressure from above.
- Move the slides to a -80 °C refrigerator, and leave them there for at least 10 min. Then take them out quickly and remove the coverslip with the help of a razor. Allow the slides to air dry at room temperature.
- Once the slides are dry add 5-30 μl of DAPI solution (vectashield with mounting medium for instance) to one of the 4 slides and cover with a new coverslip. Observe one slide under a fluorescence microscope to confirm whether the slide contains good meiotic chromosome spreads, if so the remaining slides can be used for FISH experiments. Meiotic stages can be identified using Figure 1 as a template (previously unpublished). You may seal the edges of the coverslip with regular nail polisher for short-term storage in the dark. Vectashield contains a mounting medium that allows for short term storage.
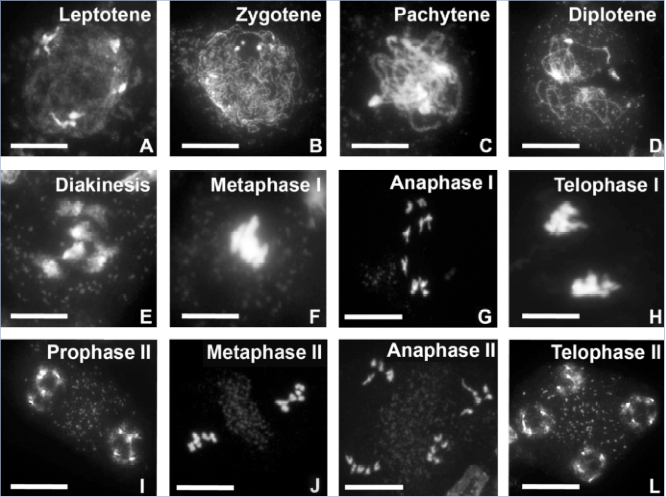
Figure 1. Stages of male meiosis in Arabidopsis as visualized by DAPI staining. In the wild type five bivalents are always observed during metaphase-I. Scale bar: A-D, 20 μm, E, F, H, 5 μm, G, I-L, 10 μm
- Prior to anther collection and fixation remove the enzyme digestion solution from the refrigerator and allow it to thaw on ice. In the meantime set a hybridization oven at 37 °C, and prepare a lunch box in which the bottom is covered by moist tissues. On the bottom of the lunch box place a rack for PCR tubes, and place the slides on top to avoid direct contact with the wet tissues. Digestion of the anthers allows free meiocytes to be released onto slides where they can be stained and observed under the microscope.
- FISH (fluorescence in situ hybridization)
- Prepare 70% formamide in 2x SSC (700 μl formamide, 100 μl 2x SSC and 100 μl distilled water). Add 50 μl to each slide, place a coverslip on top and place the slides on a hot dry block set at 80 °C. After 30 sec you may either a) dip the slide on water inside a Coplin jar to dislodge the coverslip, or b) apply 20 μl of water to lift the coverslip. Then place the slides in a Coplin jar filled with 200 ml of 70%, 90% and 100% ethanol, for 5 min each. Please notice that all ethanol solutions must be cooled in advance at -20 °C. After the bath series is finished allow the slides to air dry at room temperature.
- In the meantime prepare the pAL1 centromere probe (for this step you require a fully labeled probe). Concentration of probe: 20-30 ng/μl. Dissolve 1.5 μl in 13.5 μl of FISH buffer (for one slide), for a final FISH probe concentration of approximately 2-3 ng/μl (see Notes). Incubate the probe at 80 °C for 5-10 min to denature DNA. Spin shortly afterwards with a microcentrifuge (at approximately 10,000 rpm), at room temperature. Then quickly place the FISH probe mix on ice for at least 5 min.
- Add 15 μl of FISH probe onto each slide. Cover with a coverslip and incubate the slides on the lunch box overnight at 37 °C. The bottom of the box should contain wet paper tissues to maintain a high humidity and avoid dessication of the FISH probe.
- Next morning dip the slides on water to remove the coverslip, and then place the slides on a Coplin jar. Wash with 200 ml 2x SSC for 10 min, four times.
- You can check the condition of the slides at this point by taking one and have it stained with DAPI.
- Wash the slides on 200 ml hot 2x SSC (45 °C) for 10 min, and after that in 200 ml 1x PBS for 5 min. Washing with PBS is not considered essential and can be skipped.
- Add DAPI (5 μl) while the sample is wet. Cover with a coverslip and observe. Results can be compared with Figure 2 (previously unpublished).
- If for some reason cells are washed away, you can wash the cells on 2x SSC for 15 min once and then directly stain with DAPI.
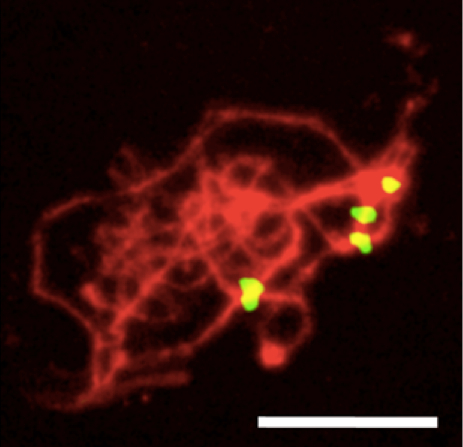
Figure 2. Arabidopsis male meiotic spread (zygotene stage) hybridized to the pAL1 centromeric 180 bp repeat (yellow) and counterstained with DAPI (red, pseudocolor). The number of centromere signals (4) roughly corresponds to the actual number of bivalents (5). Scale bar: 10 μm
- Prepare 70% formamide in 2x SSC (700 μl formamide, 100 μl 2x SSC and 100 μl distilled water). Add 50 μl to each slide, place a coverslip on top and place the slides on a hot dry block set at 80 °C. After 30 sec you may either a) dip the slide on water inside a Coplin jar to dislodge the coverslip, or b) apply 20 μl of water to lift the coverslip. Then place the slides in a Coplin jar filled with 200 ml of 70%, 90% and 100% ethanol, for 5 min each. Please notice that all ethanol solutions must be cooled in advance at -20 °C. After the bath series is finished allow the slides to air dry at room temperature.
Notes
- How to prepare a centromere probe
In order to prepare the centromere probe, PCR must be performed with specific primers (M13 forward and reverse). These two primers are required for the amplification of the Arabidopsis 180 bp centromeric repeat using the centromere-containing plasmid, pAL1 as a template. The primers are as follows:
M13-Forward: 5’-GTAAAACGACGGCCAG-3’
M13-Reverse: 5’-CAGGAAACAGCTATGAC-3’
Ordinarily, the pAL1plasmid is diluted1/100x for PCR and the resulting product is labeled for 8 h with the Fluorescein High-Prime labeling kit. Please read the manufacturer´s instructions carefully.
Recipes
- Carnoy’s solution (v/v)
95% ethanol, chloroform and glacial acetic acid (6:3:1)
- 1 L 10 mM Sodium citrate buffer
1.045 g citric acid anhydrous (MW = 192.12)
1.479 sodium citrate dihydrate (MW = 294.1)
- Enzyme digestion buffer
0.3% pectolyase Y23, 1.5 mg (0.0015 g)
0.3% cellulase, 1.5 mg (0.0015 g)
1.4 % β-glucoronidase, 7.5 mg (0.0075 g)
6% sucrose, 30 mg (0.03 g)
First dissolve enzymes in 10 mM sodium citrate (100 μl), and then continue to add sodium citrate up to 500 μl. This solution can be stored for up to one month at -20 °C.
- FISH buffer (500 μl)
10% formamide (add 50 μl 100% formamide)
2x SSC (add 50 μl 20x SSC)
10% dextran sulfate (add 100 μl 50% dextran sulfate, which is prepared by simply dissolving 1 g of dextran sulfate powder in 2 ml of distilled water, then heat to 80 °C to improve mixing)
Salmon sperm blocking DNA (add 75 μl of 25x sperm, final concentration: 250 μg/μl)
In order to prepare 25x salmon sperm DNA (25 μg/μl), dissolve 250 mg sperm powder into 1 ml of distilled water or 1x PBS
Keep at -20 °C
0.1% SDS (add 5 μl of 10% SDS)
Distilled water, 220 μl
- 10x phosphate buffered saline (PBS)
Prepare NaCl, 80 g; KCl, 2 g; Na2HPO4.7H2O, 14.4 g; KH2PO4, 2.4 g; ddH2O up to 1,000 ml
Adjust pH to 7.2-7.6
Autoclave
- 20x SSC
Prepare NaCl, 175.3 g (3 M), and sodium citrate dihydrate 88.2 g (300 mM), then dissolved in 800 ml of water
Adjust pH to 7.0
Take volume to 1,000 ml
Autoclave
Acknowledgments
This work was supported by research grants from Academia Sinica (Taiwan), the National Science and Technology Program for Agricultural Biotechnology (NSTP/AB, 098S0030055-AA), Taiwan, and the National Science Council (NSF; 99-2321-B-001-036-MY3), Taiwan, to G.-Y. Jauh; an NSF grant (MCB0718191) to C. A. Makaroff, and a study-abroad contract from the University of Costa Rica to P. Bolaños-Villegas.
References
- Bolaños-Villegas, P., Yang, X., Wang, H. J., Juan, C. T., Chuang, M. H., Makaroff, C. A. and Jauh, G. Y. (2013). Arabidopsis CHROMOSOME TRANSMISSION FIDELITY 7 (AtCTF7/ECO1) is required for DNA repair, mitosis and meiosis. Plant J 75(6): 927-9.
- Yang, X., Boateng, K. A., Yuan, L., Wu, S., Baskin, T. I. and Makaroff, C. A. (2011). The radially swollen 4 separase mutation of Arabidopsis thaliana blocks chromosome disjunction and disrupts the radial microtubule system in meiocytes. PLoS One 6(4): e19459.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Bolaños-Villegas, P., Yang, X., Makaroff, C. A. and Jauh, G. (2014). Protocol for the Preparation of Arabidopsis Meiotic Chromosome Spreads and Fluorescent in situ Hybridization. Bio-protocol 4(8): e1102. DOI: 10.21769/BioProtoc.1102.
Category
Plant Science > Plant cell biology > Cell imaging
Cell Biology > Cell structure > Chromosome
Molecular Biology > DNA > DNA structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











