- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of Haemolysin Activities in Vibrio vulnificus
Published: Vol 4, Iss 5, Mar 5, 2014 DOI: 10.21769/BioProtoc.1062 Views: 8684
Reviewed by: Fanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

H2 Production from Methyl Viologen–Dependent Hydrogenase Activity Monitored by Gas Chromatography
Nuttavut Kosem
Dec 5, 2023 1770 Views

Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1585 Views

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1928 Views
Abstract
VvhA produced by Vibrio vulnificus exhibits cytolytic activity to human cells including erythrocytes. Since haemolysis by VvhA may provide iron for bacterial growth and pathogenicity, we investigated the expression of VvhA to elucidate the regulatory roles of Fur, a major transcription factor controlling iron-homeostasis. Fur repressed the transcription of vvhBA operon via binding to the promoter region. However, haemolysin content and haemolytic activity were lowered in cell-free supernatant of fur mutant. This discrepancy between the levels of vvhA transcript and VvhA protein in fur mutant was caused by exoproteolytic activities of the elastase VvpE and another metalloprotease VvpM, which were also regulated by Fur. vvpE gene expression was repressed by Fur via binding to the Fur-box homologous region. Regulation of VvpM expression by Fur did not occur at the level of vvpM transcription. In vitro proteolysis assays showed that both proteases efficiently degraded VvhA. In addition, the extracellular levels of VvhA were higher in culture supernatants of vvpE or vvpM mutants than in the wild type. Thus this study demonstrates that Fur regulates haemolysin production at the transcription level of the vvhBA operon and at the post-translation level by regulating the expressions of two VvhA-degrading exoproteases, VvpE and VvpM.
This protocol can be applied to other Vibrio strains with haemolysin activities, such as Vibrio parahaemolyticus (V. parahaemolyticus) or other human pathogen strains with similar heamolysin activities.
Materials and Reagents
- Bacterial strains
- Wild type (Vibrio vulnificus MO6-24/O, Clinical isolate: biosafety level 2) with pRK415 plasmid (vector control)
- fur mutant with pRK415
- fur mutant with pRK415-fur
- Wild type (Vibrio vulnificus MO6-24/O, Clinical isolate: biosafety level 2) with pRK415 plasmid (vector control)
- 1% human red blood cells (RBCs) (from healthy person who is a volunteer and diluted with PBS buffer to make a 1% RBC solution)
- Tetracycline (3 mg/ml) (Sigma-Aldrich)
- PBS buffer (Sigma-Aldrich, catalog number: P5368 )
- 0.02% Triton X-100 (TritonTM X-100 for molecular biology) (Sigma-Aldrich, catalog number: T8787 )
- LB broth (see Recipes)
Equipment
- Shaking incubator
- Centrifuge (14,000 rpm or 21,000 x g)
- Spectrophotometry (540 and 600 nm)
- 0.22 micron filter
Procedure
- Grow 5 ml of bacterial cells at 30 °C shaking incubator, in modified Luria–Bertani (LB) [addition of NaCl to LB at a final concentration of 2.5% (w/v): Vibrio vulnificus is a marine bacterium, so 2.5% NaCl makes cell happy to grow.] medium supplemented with antibiotics (tetracycline 3 mg/ml).
- Take bacterial samples and measure cell density by spectrophotometry at 600 nm.
- Collect the supernatants of cultures by centrifugation (14,000 rpm, 5 min, RT) and filtration (use with syringe filter: pore size 0.22 μm).
- Prepare cell-free supernatants from each culture and serial dilute them with PBS by 1/2, 1/4 and 1/8 (final volume: 1 ml).
- Incubate 1% RBC solution with the equal volume of the diluted supernatants at 37 °C for 1 h (no agitation).
- After 1-h incubation, centrifuge of each samples to remove unlysed RBCs (14,000 rpm, 5 min, RT).
- To make the fully lysis of 1% RBC solution, use equal volume of 0.02% Triton X-100 at 37 °C for 1 h (no agitation).
- After centrifuge of each sample, take the upper part of samples. Measure the lysed RBCs samples by spectrophotometry at 540 nm, as described by Shinoda et al. (1985) (for the blank at 540 nm, you can use culture broth without bacteria cells).
- Calculate Haemolytic activity (HU) (HU was expressed as the reciprocal of the dilution factor showing 50% haemolysis) (Shinoda et al., 1985).
Representative data
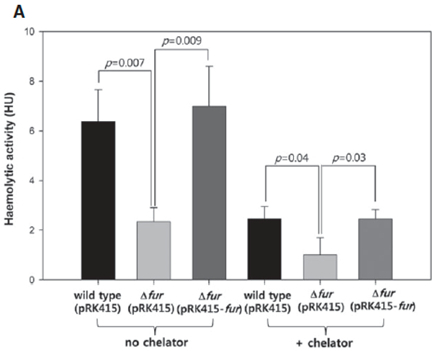
Figure 1. Haemolytic activities are decreased by fur mutation or iron deprivation. Haemolytic activities in the supernatants of wild type (pRK415), Δfur strain (pRK415) and Δfur strain (pRK415-fur) cultures. V. vulnificus strains were grown in LBS supplemented with 3 μg/ml tetracycline for 2 h, and each culture was treated for 4 h with 2,2′-dipyridyl at a concentration of 0.1 mM (+ chelator) or 0 mM (no chelator). OD600 of the cultures of wild type (pRK415) and Δfur strain (pRK415-fur) were 2.0-2.5, and OD600 of the Δfur (pRK415) cultures were 1.0-1.5. Serial dilutions of cell-free supernatants were added to 1% RBC solution, and lysed RBCs were measured by spectrophotometry. Activity was expressed as haemolytic units (HU), the reciprocal of the dilution factor showing 50% haemolysis (Lee et al., 2013).
Recipes
- LB broth
10 g of Peptone
10 g of Yeast extract
5 g Sodium Cloride per one liter
Acknowledgments
This protocol has been adapted and modified from Shinoda et al. (1985) and Lee et al. (2013). This work was supported by the Mid-Career Researcher Program through a National Research Foundation grant funded by the Ministry of Education, Science and Technology, Korea (No. 2009-0092822 to K.-H.L.) and by the Marine and Extreme Genome Research Center program of the Ministry of Land, Transport and Maritime Affair, Korea (to K.-H.L.).
References
- Lee, H. J., Kim, J. A., Lee, M. A., Park, S. J. and Lee, K. H. (2013). Regulation of haemolysin (VvhA) production by ferric uptake regulator (Fur) in Vibrio vulnificus: repression of vvhA transcription by Fur and proteolysis of VvhA by Fur-repressive exoproteases. Mol Microbiol 88(4): 813-826.
- Shinoda, S., Miyoshi, S., Yamanaka, H. and Miyoshi-Nakahara, N. (1985). Some properties of Vibrio vulnificus hemolysin. Microbiol Immunol 29(7): 583-590.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lee, H., Kim, J., Lee, M., Park, S. and Lee, K. (2014). Measurement of Haemolysin Activities in Vibrio vulnificus. Bio-protocol 4(5): e1062. DOI: 10.21769/BioProtoc.1062.
Category
Microbiology > Microbial biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Cell Biology > Cell viability > Cell lysis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









